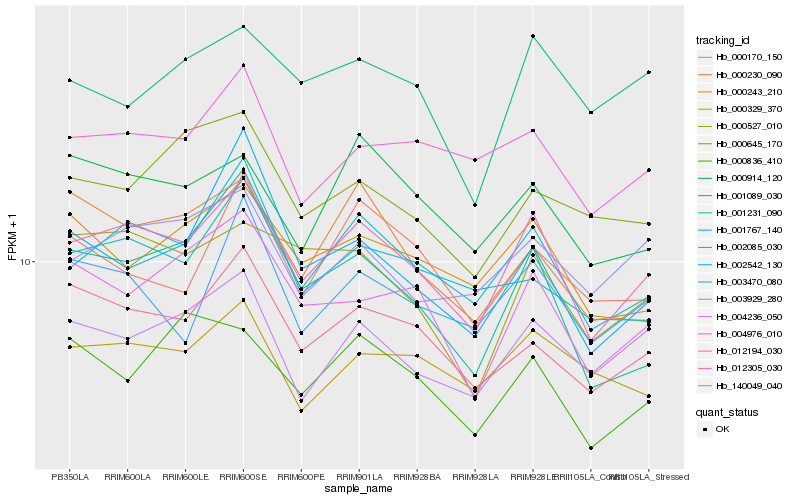
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012305_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_18979 [Jatropha curcas] |
| 2 |
Hb_002085_030 |
0.0463695946 |
- |
- |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X2 [Jatropha curcas] |
| 3 |
Hb_140049_040 |
0.0611212973 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 4 |
Hb_004976_010 |
0.0650900704 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 5 |
Hb_004236_050 |
0.0661954793 |
- |
- |
PREDICTED: nipped-B-like protein A [Jatropha curcas] |
| 6 |
Hb_000914_120 |
0.0664606006 |
- |
- |
PREDICTED: outer envelope protein 80, chloroplastic [Jatropha curcas] |
| 7 |
Hb_012194_030 |
0.0670441065 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_003929_280 |
0.0677163472 |
transcription factor |
TF Family: CAMTA |
calmodulin-binding transcription activator (camta), plants, putative [Ricinus communis] |
| 9 |
Hb_000527_010 |
0.0677233622 |
- |
- |
katanin P80 subunit, putative [Ricinus communis] |
| 10 |
Hb_003470_080 |
0.0697969772 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein PHOTOPERIOD-INDEPENDENT EARLY FLOWERING 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000230_090 |
0.0700241566 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001231_090 |
0.0707899287 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000243_210 |
0.0708133909 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_001767_140 |
0.071257335 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 15 |
Hb_000836_410 |
0.0721481529 |
- |
- |
sec10, putative [Ricinus communis] |
| 16 |
Hb_000329_370 |
0.0727942824 |
- |
- |
hypothetical protein JCGZ_14571 [Jatropha curcas] |
| 17 |
Hb_001089_030 |
0.0753649151 |
- |
- |
PREDICTED: uncharacterized protein LOC105638026 [Jatropha curcas] |
| 18 |
Hb_000170_150 |
0.0755345738 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 19 |
Hb_002542_130 |
0.0760733635 |
transcription factor |
TF Family: HB |
Homeobox protein LUMINIDEPENDENS, putative [Ricinus communis] |
| 20 |
Hb_000645_170 |
0.0760792161 |
- |
- |
conserved hypothetical protein [Ricinus communis] |