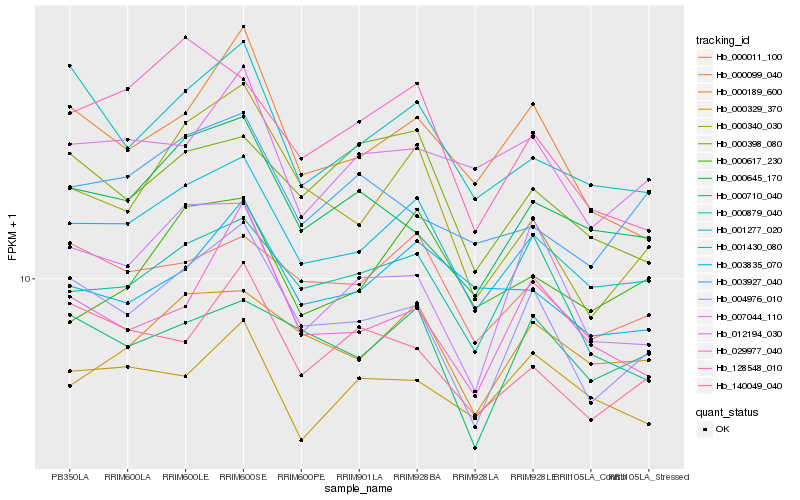
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001430_080 |
0.0 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 2 |
Hb_000011_100 |
0.0497750068 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 3 |
Hb_001277_020 |
0.0545997373 |
- |
- |
PREDICTED: armadillo repeat-containing protein LFR [Jatropha curcas] |
| 4 |
Hb_004976_010 |
0.0548707188 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 5 |
Hb_000645_170 |
0.0591197873 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003835_070 |
0.0611688942 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000879_040 |
0.0623203773 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 8 |
Hb_000189_600 |
0.0624626376 |
- |
- |
PREDICTED: protein MON2 homolog isoform X1 [Jatropha curcas] |
| 9 |
Hb_000099_040 |
0.0630160159 |
- |
- |
unnamed protein product [Coffea canephora] |
| 10 |
Hb_128548_010 |
0.0630402797 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000398_080 |
0.063217617 |
- |
- |
tip120, putative [Ricinus communis] |
| 12 |
Hb_000710_040 |
0.0637746742 |
- |
- |
PREDICTED: HEAT repeat-containing protein 6 [Jatropha curcas] |
| 13 |
Hb_140049_040 |
0.0652366914 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 14 |
Hb_000329_370 |
0.0659528065 |
- |
- |
hypothetical protein JCGZ_14571 [Jatropha curcas] |
| 15 |
Hb_012194_030 |
0.0660247763 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000617_230 |
0.0663711104 |
- |
- |
PREDICTED: tafazzin isoform X1 [Jatropha curcas] |
| 17 |
Hb_029977_040 |
0.0674855338 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X4 [Jatropha curcas] |
| 18 |
Hb_000340_030 |
0.067829315 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 19 |
Hb_003927_040 |
0.0685633525 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 20 |
Hb_007044_110 |
0.0696371561 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |