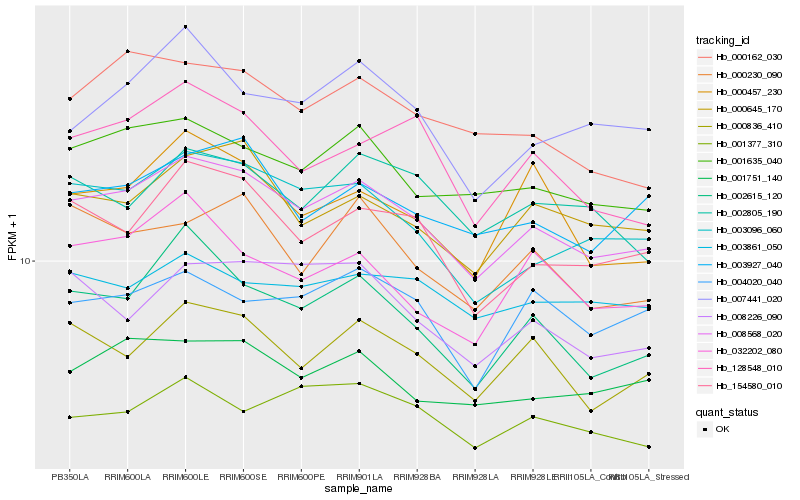
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008568_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 2 |
Hb_003096_060 |
0.0513160672 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 3 |
Hb_001635_040 |
0.0518732748 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 4 |
Hb_154580_010 |
0.0526638789 |
- |
- |
hypothetical protein PRUPE_ppa002109mg [Prunus persica] |
| 5 |
Hb_000836_410 |
0.0548711718 |
- |
- |
sec10, putative [Ricinus communis] |
| 6 |
Hb_000645_170 |
0.0562654572 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002615_120 |
0.0573790454 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001751_140 |
0.0579531418 |
- |
- |
PREDICTED: uncharacterized protein LOC105648510 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003927_040 |
0.0610039582 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 10 |
Hb_000162_030 |
0.0640433424 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_128548_010 |
0.0648944842 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007441_020 |
0.0653888017 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 13 |
Hb_008226_090 |
0.0661904374 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002805_190 |
0.0665608853 |
- |
- |
spliceosome associated protein, putative [Ricinus communis] |
| 15 |
Hb_001377_310 |
0.066620107 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 16 |
Hb_032202_080 |
0.0668294682 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000230_090 |
0.0673188015 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004020_040 |
0.0674242791 |
- |
- |
PREDICTED: GTP-binding protein At3g49725, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000457_230 |
0.0674750474 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_003861_050 |
0.0676397672 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |