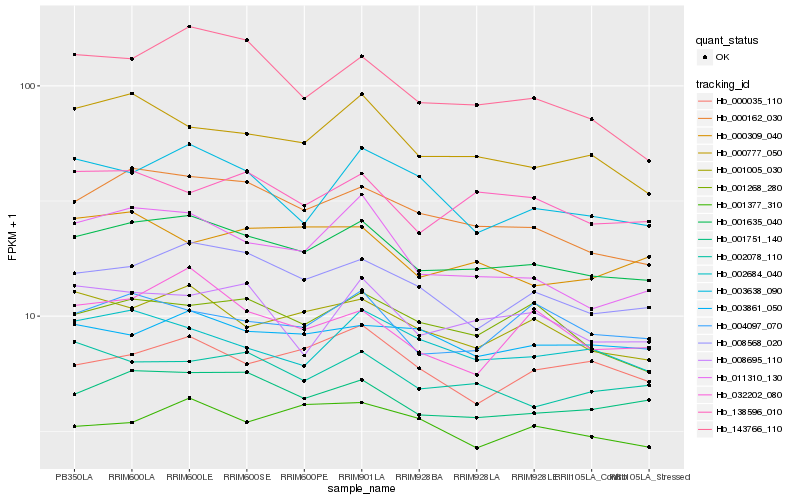
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001635_040 |
0.0 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 2 |
Hb_001751_140 |
0.0475523177 |
- |
- |
PREDICTED: uncharacterized protein LOC105648510 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000162_030 |
0.0493110898 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_008568_020 |
0.0518732748 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 5 |
Hb_001005_030 |
0.0577531038 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 6 |
Hb_000777_050 |
0.0639156141 |
- |
- |
hypothetical protein CICLE_v10008422mg [Citrus clementina] |
| 7 |
Hb_138596_010 |
0.0654263702 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g13690 [Jatropha curcas] |
| 8 |
Hb_002078_110 |
0.065558005 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 9 |
Hb_002684_040 |
0.0659972453 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001268_280 |
0.0666474006 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_011310_130 |
0.0669599854 |
- |
- |
DEAD/DEAH box RNA helicase family protein isoform 1 [Theobroma cacao] |
| 12 |
Hb_003861_050 |
0.0677913322 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 13 |
Hb_004097_070 |
0.0680833435 |
- |
- |
PREDICTED: F-box protein At2g26160-like [Jatropha curcas] |
| 14 |
Hb_008695_110 |
0.0717392452 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105638874 isoform X1 [Jatropha curcas] |
| 15 |
Hb_143766_110 |
0.0734949068 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 1 [Jatropha curcas] |
| 16 |
Hb_003638_090 |
0.07387586 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45a-like [Jatropha curcas] |
| 17 |
Hb_000035_110 |
0.0739875349 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 5 [Jatropha curcas] |
| 18 |
Hb_000309_040 |
0.074280835 |
- |
- |
PREDICTED: protein farnesyltransferase subunit beta isoform X1 [Jatropha curcas] |
| 19 |
Hb_001377_310 |
0.0743198682 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 20 |
Hb_032202_080 |
0.0745014785 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |