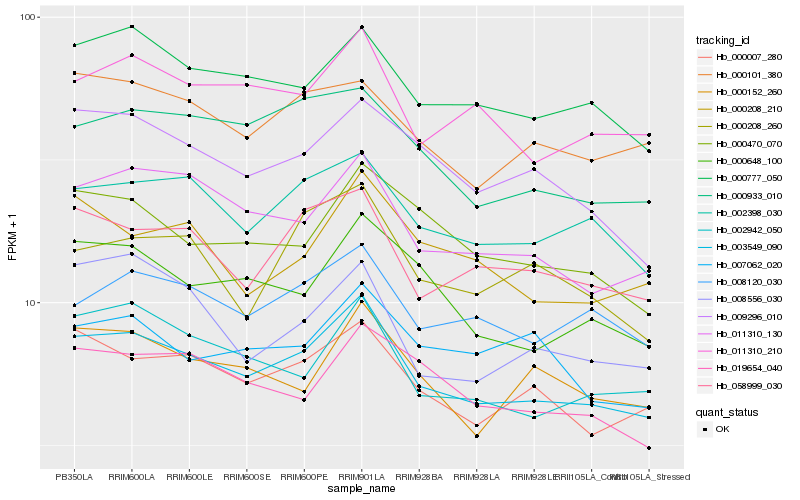
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003549_090 |
0.0 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 6 [Jatropha curcas] |
| 2 |
Hb_002398_030 |
0.0607202687 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 3 |
Hb_011310_130 |
0.0611532034 |
- |
- |
DEAD/DEAH box RNA helicase family protein isoform 1 [Theobroma cacao] |
| 4 |
Hb_000777_050 |
0.061294323 |
- |
- |
hypothetical protein CICLE_v10008422mg [Citrus clementina] |
| 5 |
Hb_000007_280 |
0.0614320082 |
- |
- |
hypothetical protein VITISV_000066 [Vitis vinifera] |
| 6 |
Hb_011310_210 |
0.0666814938 |
- |
- |
PREDICTED: serine/threonine-protein kinase PRP4 homolog [Jatropha curcas] |
| 7 |
Hb_058999_030 |
0.066961321 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 8 |
Hb_000933_010 |
0.0686750307 |
- |
- |
- |
| 9 |
Hb_008120_030 |
0.0690500685 |
- |
- |
PREDICTED: uncharacterized protein LOC101216675 [Cucumis sativus] |
| 10 |
Hb_000648_100 |
0.0693512838 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 11 |
Hb_002942_050 |
0.0704631443 |
- |
- |
PREDICTED: APO protein 3, mitochondrial [Jatropha curcas] |
| 12 |
Hb_009296_010 |
0.0716688799 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor GTP-binding subunit ERF3A isoform X3 [Jatropha curcas] |
| 13 |
Hb_000101_380 |
0.0738949666 |
- |
- |
PREDICTED: clathrin light chain 2-like isoform X2 [Populus euphratica] |
| 14 |
Hb_000470_070 |
0.0743213336 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP43-like [Jatropha curcas] |
| 15 |
Hb_019654_040 |
0.0762674065 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase Pus10 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000152_260 |
0.0769044988 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 17 |
Hb_000208_210 |
0.0797279774 |
- |
- |
PREDICTED: RNA-binding protein Nova-2 [Jatropha curcas] |
| 18 |
Hb_000208_260 |
0.0810418515 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4 [Jatropha curcas] |
| 19 |
Hb_007062_020 |
0.0843700275 |
- |
- |
gcn4-complementing protein, putative [Ricinus communis] |
| 20 |
Hb_008556_030 |
0.0849355301 |
- |
- |
PREDICTED: uncharacterized protein LOC105648021 isoform X1 [Jatropha curcas] |