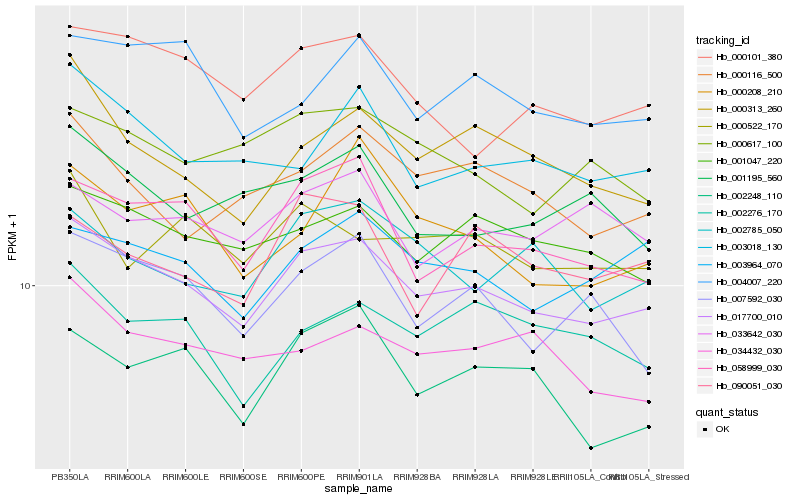
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017700_010 |
0.0 |
- |
- |
PREDICTED: probable protein S-acyltransferase 15 [Jatropha curcas] |
| 2 |
Hb_058999_030 |
0.0650124277 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 3 |
Hb_001047_220 |
0.0651930262 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 4 |
Hb_003964_070 |
0.0695022107 |
- |
- |
hypothetical protein JCGZ_15695 [Jatropha curcas] |
| 5 |
Hb_000101_380 |
0.0711771422 |
- |
- |
PREDICTED: clathrin light chain 2-like isoform X2 [Populus euphratica] |
| 6 |
Hb_000313_260 |
0.0722288899 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 7 |
Hb_003018_130 |
0.0730857327 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 8 |
Hb_002785_050 |
0.0746281715 |
- |
- |
PREDICTED: adenylosuccinate lyase-like [Jatropha curcas] |
| 9 |
Hb_004007_220 |
0.0750603985 |
- |
- |
PREDICTED: casein kinase II subunit alpha [Jatropha curcas] |
| 10 |
Hb_007592_030 |
0.0752329853 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 11 |
Hb_000116_500 |
0.0754407111 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 11 [Jatropha curcas] |
| 12 |
Hb_000617_100 |
0.0763694928 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 13 |
Hb_034432_030 |
0.077428996 |
- |
- |
PREDICTED: probable glycosyltransferase At5g20260 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001195_560 |
0.0792243085 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002276_170 |
0.0796897069 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_033642_030 |
0.0800370205 |
- |
- |
PREDICTED: G-protein coupled receptor 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000522_170 |
0.0812247191 |
- |
- |
PREDICTED: phosphatidylinositol:ceramide inositolphosphotransferase 1 [Jatropha curcas] |
| 18 |
Hb_002248_110 |
0.0827809713 |
- |
- |
hypothetical protein POPTR_0001s41050g [Populus trichocarpa] |
| 19 |
Hb_090051_030 |
0.0830977976 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 20 |
Hb_000208_210 |
0.0831302974 |
- |
- |
PREDICTED: RNA-binding protein Nova-2 [Jatropha curcas] |