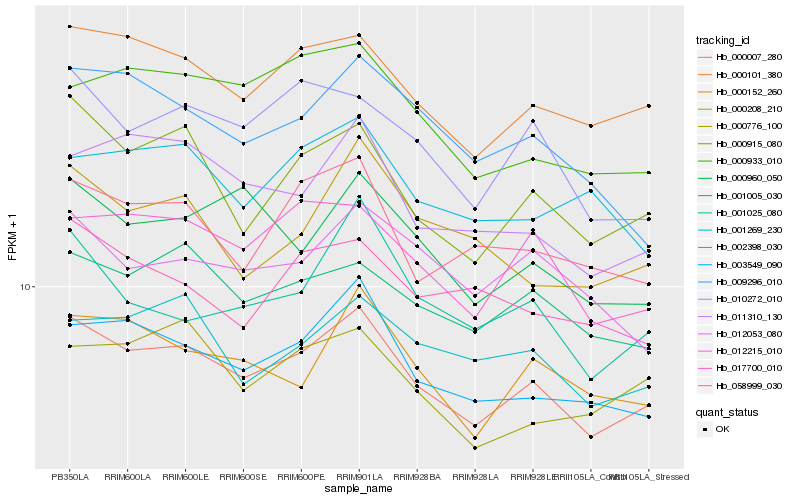
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000007_280 |
0.0 |
- |
- |
hypothetical protein VITISV_000066 [Vitis vinifera] |
| 2 |
Hb_000101_380 |
0.053717091 |
- |
- |
PREDICTED: clathrin light chain 2-like isoform X2 [Populus euphratica] |
| 3 |
Hb_003549_090 |
0.0614320082 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 6 [Jatropha curcas] |
| 4 |
Hb_000915_080 |
0.0628084703 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 5 |
Hb_001005_030 |
0.0680145759 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 6 |
Hb_011310_130 |
0.0713072403 |
- |
- |
DEAD/DEAH box RNA helicase family protein isoform 1 [Theobroma cacao] |
| 7 |
Hb_010272_010 |
0.0718699847 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 8 |
Hb_000152_260 |
0.0726029772 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 9 |
Hb_058999_030 |
0.0749015145 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 10 |
Hb_000933_010 |
0.076305771 |
- |
- |
- |
| 11 |
Hb_012215_010 |
0.0776318805 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 12 |
Hb_000960_050 |
0.0781166153 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 13 |
Hb_001269_230 |
0.0782250373 |
- |
- |
PREDICTED: tRNA-splicing endonuclease subunit Sen54 [Jatropha curcas] |
| 14 |
Hb_009296_010 |
0.0797175777 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor GTP-binding subunit ERF3A isoform X3 [Jatropha curcas] |
| 15 |
Hb_001025_080 |
0.0810860409 |
- |
- |
hypothetical protein JCGZ_11431 [Jatropha curcas] |
| 16 |
Hb_000776_100 |
0.0818531739 |
- |
- |
Cleavage stimulation factor 50 kDa subunit, putative [Ricinus communis] |
| 17 |
Hb_017700_010 |
0.0832126398 |
- |
- |
PREDICTED: probable protein S-acyltransferase 15 [Jatropha curcas] |
| 18 |
Hb_012053_080 |
0.0841102289 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 19 |
Hb_000208_210 |
0.0846690591 |
- |
- |
PREDICTED: RNA-binding protein Nova-2 [Jatropha curcas] |
| 20 |
Hb_002398_030 |
0.0849936949 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |