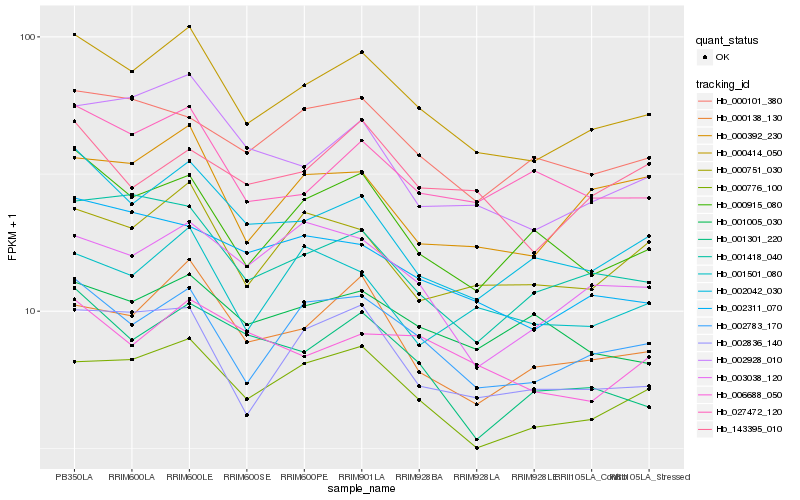
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000414_050 |
0.0 |
- |
- |
hypothetical protein JCGZ_19588 [Jatropha curcas] |
| 2 |
Hb_002783_170 |
0.0519957456 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X2 [Jatropha curcas] |
| 3 |
Hb_000776_100 |
0.0582114663 |
- |
- |
Cleavage stimulation factor 50 kDa subunit, putative [Ricinus communis] |
| 4 |
Hb_002042_030 |
0.067107663 |
- |
- |
PREDICTED: protein YIPF1 homolog [Jatropha curcas] |
| 5 |
Hb_000138_130 |
0.0683282634 |
- |
- |
hypothetical protein JCGZ_25540 [Jatropha curcas] |
| 6 |
Hb_002836_140 |
0.0710364154 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 7 |
Hb_000915_080 |
0.073540958 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 8 |
Hb_000751_030 |
0.0760141701 |
- |
- |
Vesicle transport protein GOT1B, putative [Ricinus communis] |
| 9 |
Hb_002928_010 |
0.0770245221 |
- |
- |
PREDICTED: WD repeat-containing protein DWA2 [Jatropha curcas] |
| 10 |
Hb_001418_040 |
0.0771287436 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1-like [Jatropha curcas] |
| 11 |
Hb_027472_120 |
0.0779598299 |
- |
- |
PREDICTED: thiosulfate/3-mercaptopyruvate sulfurtransferase 1, mitochondrial isoform X1 [Jatropha curcas] |
| 12 |
Hb_000392_230 |
0.0805051719 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002311_070 |
0.0810388691 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 14 |
Hb_000101_380 |
0.0828478336 |
- |
- |
PREDICTED: clathrin light chain 2-like isoform X2 [Populus euphratica] |
| 15 |
Hb_001301_220 |
0.0829258073 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 4 [Jatropha curcas] |
| 16 |
Hb_003038_120 |
0.0841651029 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 17 |
Hb_001501_080 |
0.087357014 |
- |
- |
PREDICTED: uncharacterized protein LOC105640832 isoform X1 [Jatropha curcas] |
| 18 |
Hb_006688_050 |
0.0878087851 |
- |
- |
PREDICTED: cytochrome c oxidase assembly protein COX15 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001005_030 |
0.0892477865 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 20 |
Hb_143395_010 |
0.0897919953 |
- |
- |
PREDICTED: uncharacterized protein LOC105642243 [Jatropha curcas] |