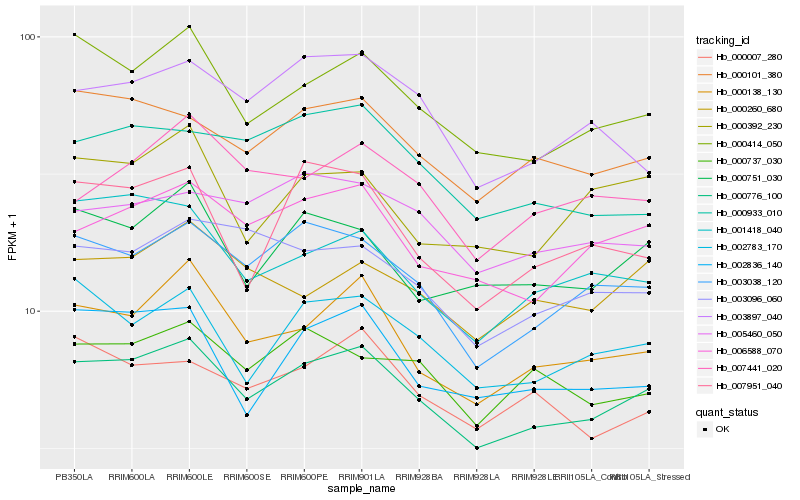
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000776_100 |
0.0 |
- |
- |
Cleavage stimulation factor 50 kDa subunit, putative [Ricinus communis] |
| 2 |
Hb_000414_050 |
0.0582114663 |
- |
- |
hypothetical protein JCGZ_19588 [Jatropha curcas] |
| 3 |
Hb_003038_120 |
0.0586149506 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 4 |
Hb_000138_130 |
0.0609769387 |
- |
- |
hypothetical protein JCGZ_25540 [Jatropha curcas] |
| 5 |
Hb_006588_070 |
0.0620534661 |
- |
- |
20 kD nuclear cap binding protein, putative [Ricinus communis] |
| 6 |
Hb_000101_380 |
0.0633414266 |
- |
- |
PREDICTED: clathrin light chain 2-like isoform X2 [Populus euphratica] |
| 7 |
Hb_002783_170 |
0.0679063685 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X2 [Jatropha curcas] |
| 8 |
Hb_000751_030 |
0.0728184054 |
- |
- |
Vesicle transport protein GOT1B, putative [Ricinus communis] |
| 9 |
Hb_002836_140 |
0.07740454 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 10 |
Hb_003096_060 |
0.078840907 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 11 |
Hb_005460_050 |
0.079621188 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 12 |
Hb_000737_030 |
0.0798609538 |
- |
- |
PREDICTED: probable galacturonosyltransferase 3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000260_680 |
0.0803103509 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 14 |
Hb_001418_040 |
0.0805604949 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1-like [Jatropha curcas] |
| 15 |
Hb_007441_020 |
0.0810021296 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 16 |
Hb_000933_010 |
0.0811828305 |
- |
- |
- |
| 17 |
Hb_007951_040 |
0.0812822233 |
- |
- |
signal peptidase family protein [Populus trichocarpa] |
| 18 |
Hb_000392_230 |
0.0814717691 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000007_280 |
0.0818531739 |
- |
- |
hypothetical protein VITISV_000066 [Vitis vinifera] |
| 20 |
Hb_003897_040 |
0.0818557253 |
- |
- |
translocon-associated protein, beta subunit precursor, putative [Ricinus communis] |