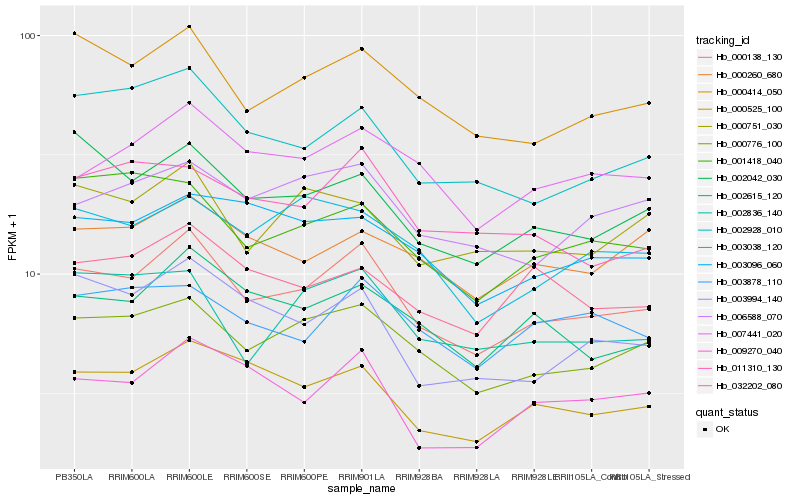
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000138_130 |
0.0 |
- |
- |
hypothetical protein JCGZ_25540 [Jatropha curcas] |
| 2 |
Hb_000776_100 |
0.0609769387 |
- |
- |
Cleavage stimulation factor 50 kDa subunit, putative [Ricinus communis] |
| 3 |
Hb_000414_050 |
0.0683282634 |
- |
- |
hypothetical protein JCGZ_19588 [Jatropha curcas] |
| 4 |
Hb_000525_100 |
0.0783949687 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 5 |
Hb_002615_120 |
0.0808108775 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002928_010 |
0.0822011451 |
- |
- |
PREDICTED: WD repeat-containing protein DWA2 [Jatropha curcas] |
| 7 |
Hb_002042_030 |
0.0827708569 |
- |
- |
PREDICTED: protein YIPF1 homolog [Jatropha curcas] |
| 8 |
Hb_007441_020 |
0.0828133156 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 9 |
Hb_006588_070 |
0.0856838446 |
- |
- |
20 kD nuclear cap binding protein, putative [Ricinus communis] |
| 10 |
Hb_003994_140 |
0.0877582657 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_009270_040 |
0.0879467463 |
- |
- |
- |
| 12 |
Hb_000751_030 |
0.0899850396 |
- |
- |
Vesicle transport protein GOT1B, putative [Ricinus communis] |
| 13 |
Hb_003878_110 |
0.0901773405 |
- |
- |
PREDICTED: uncharacterized protein LOC105637024 [Jatropha curcas] |
| 14 |
Hb_003038_120 |
0.0903453477 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 15 |
Hb_002836_140 |
0.090853554 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 16 |
Hb_000260_680 |
0.0911615728 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 17 |
Hb_003096_060 |
0.0918633204 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 18 |
Hb_032202_080 |
0.092033721 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_011310_130 |
0.0923270261 |
- |
- |
DEAD/DEAH box RNA helicase family protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_001418_040 |
0.092342862 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1-like [Jatropha curcas] |