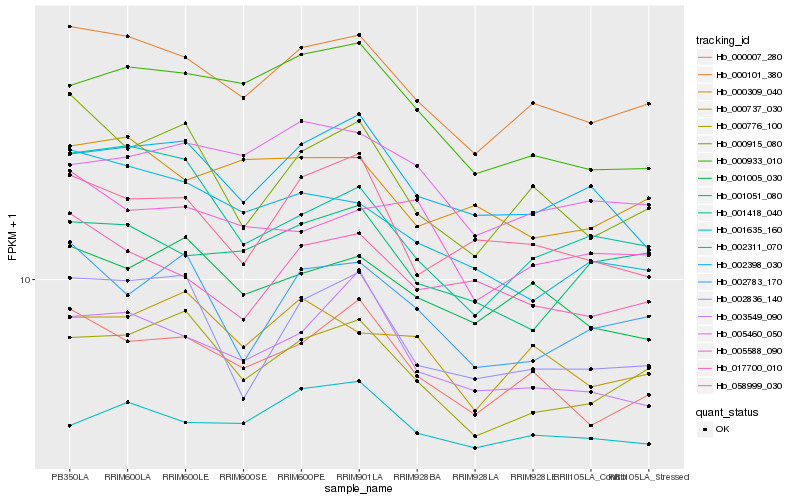
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000101_380 |
0.0 |
- |
- |
PREDICTED: clathrin light chain 2-like isoform X2 [Populus euphratica] |
| 2 |
Hb_000007_280 |
0.053717091 |
- |
- |
hypothetical protein VITISV_000066 [Vitis vinifera] |
| 3 |
Hb_000776_100 |
0.0633414266 |
- |
- |
Cleavage stimulation factor 50 kDa subunit, putative [Ricinus communis] |
| 4 |
Hb_000915_080 |
0.067135195 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 5 |
Hb_001005_030 |
0.0678713522 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 6 |
Hb_017700_010 |
0.0711771422 |
- |
- |
PREDICTED: probable protein S-acyltransferase 15 [Jatropha curcas] |
| 7 |
Hb_000737_030 |
0.0733050571 |
- |
- |
PREDICTED: probable galacturonosyltransferase 3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002398_030 |
0.0733217158 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_003549_090 |
0.0738949666 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 6 [Jatropha curcas] |
| 10 |
Hb_001051_080 |
0.0746058193 |
- |
- |
hypothetical protein EUGRSUZ_B02508 [Eucalyptus grandis] |
| 11 |
Hb_000309_040 |
0.074770634 |
- |
- |
PREDICTED: protein farnesyltransferase subunit beta isoform X1 [Jatropha curcas] |
| 12 |
Hb_002311_070 |
0.07538863 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 13 |
Hb_058999_030 |
0.0755191593 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 14 |
Hb_000933_010 |
0.0756299231 |
- |
- |
- |
| 15 |
Hb_001418_040 |
0.0759836431 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1-like [Jatropha curcas] |
| 16 |
Hb_005588_090 |
0.0761545551 |
- |
- |
hypothetical protein CICLE_v10003480mg [Citrus clementina] |
| 17 |
Hb_005460_050 |
0.0770072537 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 18 |
Hb_002783_170 |
0.0770452837 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X2 [Jatropha curcas] |
| 19 |
Hb_002836_140 |
0.0772669941 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 20 |
Hb_001635_160 |
0.0779614091 |
- |
- |
hypothetical protein JCGZ_24810 [Jatropha curcas] |