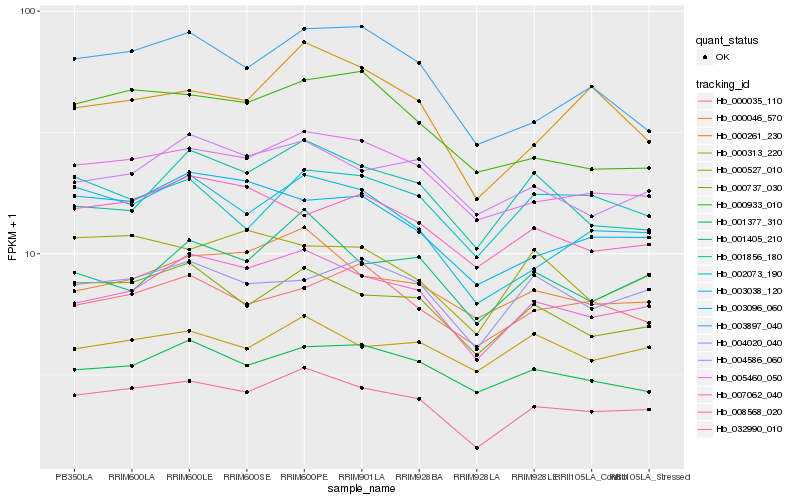
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032990_010 |
0.0 |
- |
- |
PREDICTED: fanconi-associated nuclease 1 homolog [Jatropha curcas] |
| 2 |
Hb_007062_040 |
0.045192145 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 3 |
Hb_005460_050 |
0.0524243443 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 4 |
Hb_000737_030 |
0.0565943121 |
- |
- |
PREDICTED: probable galacturonosyltransferase 3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003038_120 |
0.0658858607 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 6 |
Hb_000046_570 |
0.0674936793 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 7 |
Hb_004586_060 |
0.072222191 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 8 |
Hb_004020_040 |
0.072318907 |
- |
- |
PREDICTED: GTP-binding protein At3g49725, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001405_210 |
0.0734171028 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 10 |
Hb_001377_310 |
0.0735104668 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 11 |
Hb_000261_230 |
0.0759725334 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Vitis vinifera] |
| 12 |
Hb_003897_040 |
0.0767987237 |
- |
- |
translocon-associated protein, beta subunit precursor, putative [Ricinus communis] |
| 13 |
Hb_003096_060 |
0.0778298595 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 14 |
Hb_000035_110 |
0.0783478494 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 5 [Jatropha curcas] |
| 15 |
Hb_000313_220 |
0.079470871 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000527_010 |
0.0802547379 |
- |
- |
katanin P80 subunit, putative [Ricinus communis] |
| 17 |
Hb_002073_190 |
0.0805023128 |
- |
- |
PREDICTED: uncharacterized protein LOC105649812 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000933_010 |
0.0824191405 |
- |
- |
- |
| 19 |
Hb_001856_180 |
0.082464962 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 20 |
Hb_008568_020 |
0.0842516607 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |