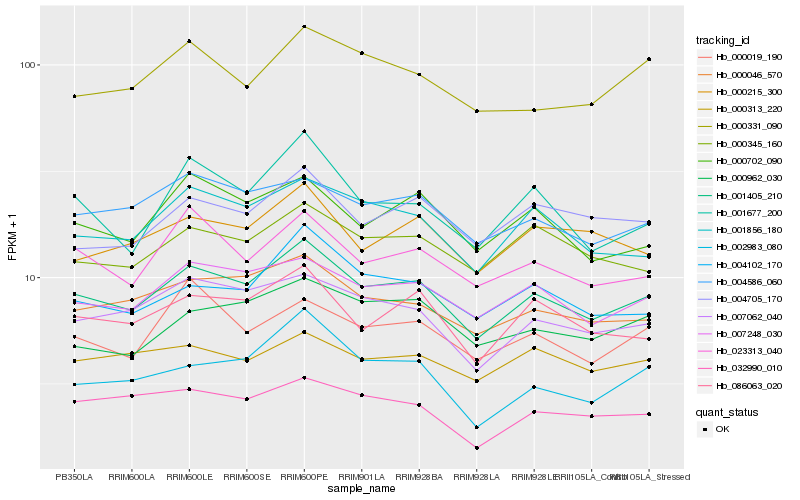
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001405_210 |
0.0 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 2 |
Hb_007248_030 |
0.0535001082 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 3 |
Hb_000046_570 |
0.059083803 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 4 |
Hb_007062_040 |
0.0604712223 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 5 |
Hb_001856_180 |
0.0622076071 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 6 |
Hb_086063_020 |
0.0626085878 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 [Jatropha curcas] |
| 7 |
Hb_004586_060 |
0.0637646407 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 8 |
Hb_004102_170 |
0.0646268997 |
- |
- |
PREDICTED: lys-63-specific deubiquitinase BRCC36-like [Jatropha curcas] |
| 9 |
Hb_023313_040 |
0.0652107845 |
- |
- |
PREDICTED: uncharacterized protein LOC105640827 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000345_160 |
0.0680188906 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 11 |
Hb_000702_090 |
0.0681157476 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 12 |
Hb_001677_200 |
0.0719979576 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004705_170 |
0.0726808807 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 14 |
Hb_032990_010 |
0.0734171028 |
- |
- |
PREDICTED: fanconi-associated nuclease 1 homolog [Jatropha curcas] |
| 15 |
Hb_000215_300 |
0.0735898666 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 16 |
Hb_000962_030 |
0.0738091562 |
- |
- |
PREDICTED: metal tolerance protein C2 [Jatropha curcas] |
| 17 |
Hb_000313_220 |
0.0743934585 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000331_090 |
0.0745624275 |
- |
- |
hypothetical protein PRUPE_ppa010923mg [Prunus persica] |
| 19 |
Hb_000019_190 |
0.0748415935 |
- |
- |
hypothetical protein POPTR_0002s23900g [Populus trichocarpa] |
| 20 |
Hb_002983_080 |
0.0756023889 |
- |
- |
acetylornithine aminotransferase, partial [Prunus persica] |