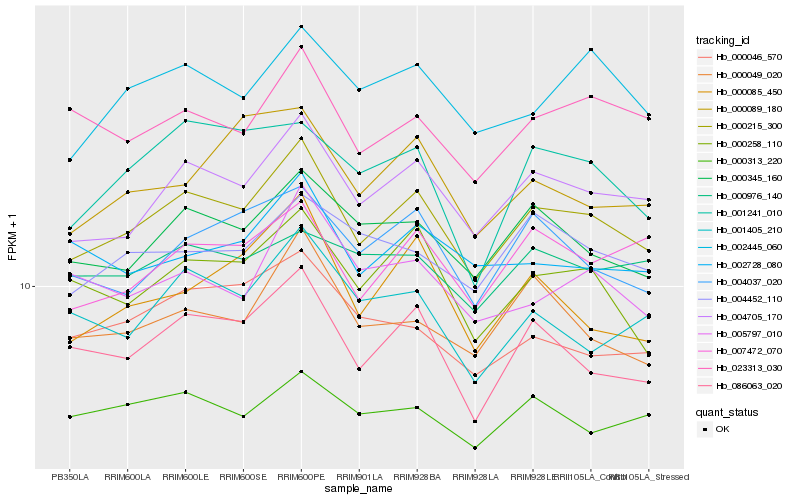
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000215_300 |
0.0 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 2 |
Hb_004705_170 |
0.0399865892 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 3 |
Hb_086063_020 |
0.0562697478 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 [Jatropha curcas] |
| 4 |
Hb_000345_160 |
0.057229945 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 5 |
Hb_000258_110 |
0.0637330559 |
- |
- |
PREDICTED: probable apyrase 6 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004037_020 |
0.064546866 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |
| 7 |
Hb_007472_070 |
0.0662692517 |
- |
- |
cir, putative [Ricinus communis] |
| 8 |
Hb_002445_060 |
0.0666852375 |
- |
- |
arf gtpase-activating protein, putative [Ricinus communis] |
| 9 |
Hb_000049_020 |
0.0674480404 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 10 |
Hb_023313_030 |
0.0704375674 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000085_450 |
0.0722025822 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 12 |
Hb_004452_110 |
0.0727814095 |
- |
- |
PREDICTED: uncharacterized protein LOC105639575 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001241_010 |
0.0729755748 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_001405_210 |
0.0735898666 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 15 |
Hb_000313_220 |
0.074131688 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000046_570 |
0.0745301254 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 17 |
Hb_002728_080 |
0.0748263094 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 18 |
Hb_000089_180 |
0.0748575356 |
- |
- |
PREDICTED: homoserine kinase-like [Jatropha curcas] |
| 19 |
Hb_005797_010 |
0.0761183086 |
- |
- |
PREDICTED: lysophospholipid acyltransferase 1-like [Jatropha curcas] |
| 20 |
Hb_000976_140 |
0.0780133709 |
- |
- |
PREDICTED: uncharacterized protein LOC105646208 isoform X1 [Jatropha curcas] |