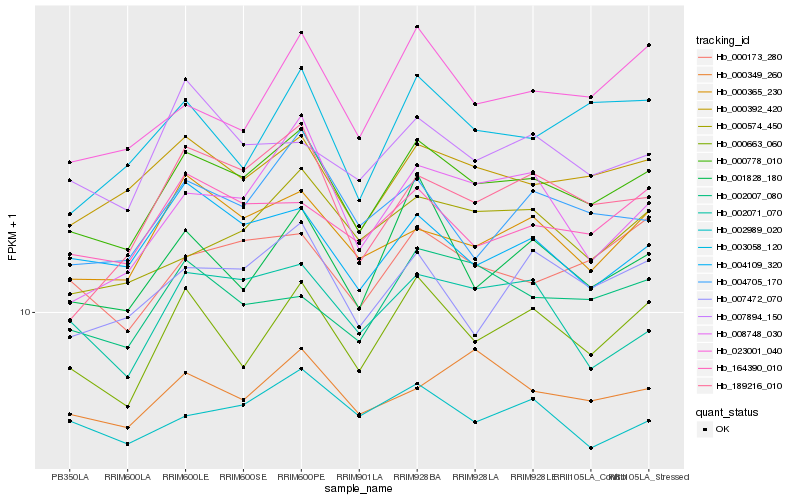
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000778_010 |
0.0 |
- |
- |
hypothetical protein [Bacillus subtilis] |
| 2 |
Hb_000392_420 |
0.0513191207 |
- |
- |
PREDICTED: probable aldehyde dehydrogenase isoform X1 [Jatropha curcas] |
| 3 |
Hb_000663_060 |
0.0520334635 |
- |
- |
hypothetical protein JCGZ_16277 [Jatropha curcas] |
| 4 |
Hb_000365_230 |
0.0581492828 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |
| 5 |
Hb_164390_010 |
0.0589404626 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 6 |
Hb_007472_070 |
0.0624055943 |
- |
- |
cir, putative [Ricinus communis] |
| 7 |
Hb_000173_280 |
0.0630200347 |
- |
- |
Speckle-type POZ protein, putative [Ricinus communis] |
| 8 |
Hb_004705_170 |
0.0641906424 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 9 |
Hb_001828_180 |
0.0647518266 |
- |
- |
26S proteasome regulatory subunit S3, putative [Ricinus communis] |
| 10 |
Hb_004109_320 |
0.0652151525 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 52 A [Jatropha curcas] |
| 11 |
Hb_002007_080 |
0.0661480648 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 12 |
Hb_002071_070 |
0.0676416794 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007894_150 |
0.067864946 |
- |
- |
hypothetical protein EUGRSUZ_F03462 [Eucalyptus grandis] |
| 14 |
Hb_002989_020 |
0.068238179 |
- |
- |
PREDICTED: uncharacterized protein LOC105630013 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000574_450 |
0.0682400226 |
- |
- |
PREDICTED: signal recognition particle 54 kDa protein 2 [Jatropha curcas] |
| 16 |
Hb_000349_260 |
0.0689557146 |
- |
- |
PREDICTED: transmembrane protein 64 [Jatropha curcas] |
| 17 |
Hb_189216_010 |
0.0698175754 |
- |
- |
PREDICTED: uncharacterized protein LOC105642236 isoform X1 [Jatropha curcas] |
| 18 |
Hb_008748_030 |
0.0702803041 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_023001_040 |
0.0705271238 |
- |
- |
Metallopeptidase M24 family protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_003058_120 |
0.0706846607 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 4 homolog [Jatropha curcas] |