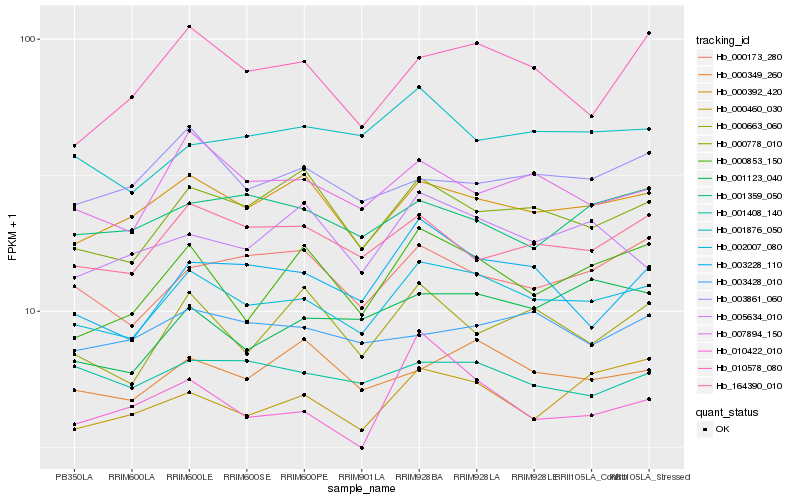
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002007_080 |
0.0 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 2 |
Hb_000392_420 |
0.0587386639 |
- |
- |
PREDICTED: probable aldehyde dehydrogenase isoform X1 [Jatropha curcas] |
| 3 |
Hb_000778_010 |
0.0661480648 |
- |
- |
hypothetical protein [Bacillus subtilis] |
| 4 |
Hb_007894_150 |
0.0662443583 |
- |
- |
hypothetical protein EUGRSUZ_F03462 [Eucalyptus grandis] |
| 5 |
Hb_164390_010 |
0.0685901136 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 6 |
Hb_000349_260 |
0.0708802401 |
- |
- |
PREDICTED: transmembrane protein 64 [Jatropha curcas] |
| 7 |
Hb_000173_280 |
0.0715175777 |
- |
- |
Speckle-type POZ protein, putative [Ricinus communis] |
| 8 |
Hb_003228_110 |
0.0723171838 |
- |
- |
PREDICTED: uncharacterized protein LOC105637245 [Jatropha curcas] |
| 9 |
Hb_000460_030 |
0.0744294478 |
- |
- |
4-hydroxybenzoate octaprenyltransferase, putative [Ricinus communis] |
| 10 |
Hb_001876_050 |
0.0745091951 |
- |
- |
hypothetical protein JCGZ_01286 [Jatropha curcas] |
| 11 |
Hb_000853_150 |
0.076055066 |
- |
- |
Fumarase 1 isoform 2 [Theobroma cacao] |
| 12 |
Hb_001408_140 |
0.0781424931 |
- |
- |
hypothetical protein JCGZ_00234 [Jatropha curcas] |
| 13 |
Hb_003861_060 |
0.078882275 |
- |
- |
PREDICTED: treacle protein [Jatropha curcas] |
| 14 |
Hb_010578_080 |
0.0792271486 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA16-like [Populus euphratica] |
| 15 |
Hb_003428_010 |
0.0793425282 |
- |
- |
PREDICTED: probable NAD(P)H dehydrogenase (quinone) FQR1-like 1 [Jatropha curcas] |
| 16 |
Hb_000663_060 |
0.0794317997 |
- |
- |
hypothetical protein JCGZ_16277 [Jatropha curcas] |
| 17 |
Hb_001359_050 |
0.0806096047 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001123_040 |
0.0813721431 |
- |
- |
PREDICTED: R3H domain-containing protein 1-like [Jatropha curcas] |
| 19 |
Hb_005634_010 |
0.0824438316 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 20 |
Hb_010422_010 |
0.082544645 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |