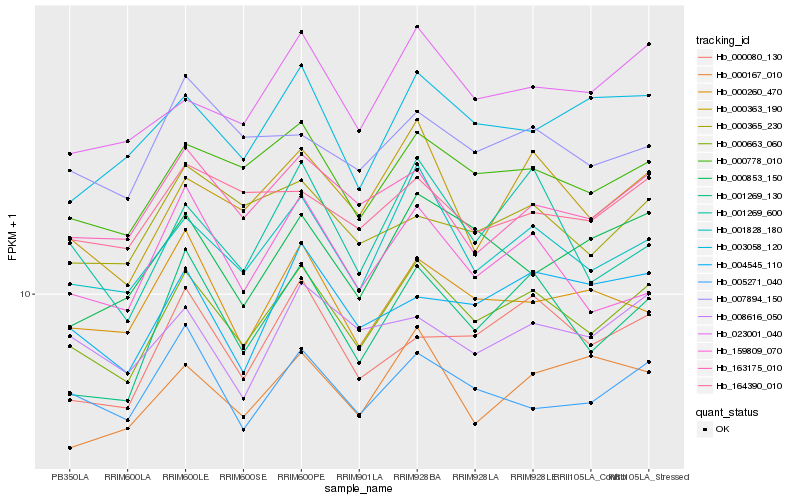
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000663_060 |
0.0 |
- |
- |
hypothetical protein JCGZ_16277 [Jatropha curcas] |
| 2 |
Hb_001828_180 |
0.0508118455 |
- |
- |
26S proteasome regulatory subunit S3, putative [Ricinus communis] |
| 3 |
Hb_000778_010 |
0.0520334635 |
- |
- |
hypothetical protein [Bacillus subtilis] |
| 4 |
Hb_001269_130 |
0.0600866493 |
- |
- |
plant poly(A)+ RNA export protein, putative [Ricinus communis] |
| 5 |
Hb_000363_190 |
0.0630809686 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-2-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_023001_040 |
0.0679661419 |
- |
- |
Metallopeptidase M24 family protein isoform 1 [Theobroma cacao] |
| 7 |
Hb_163175_010 |
0.0700327559 |
- |
- |
hypothetical protein CISIN_1g036035mg, partial [Citrus sinensis] |
| 8 |
Hb_005271_040 |
0.0709184014 |
- |
- |
PREDICTED: serine racemase [Jatropha curcas] |
| 9 |
Hb_000080_130 |
0.0719662027 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 10 |
Hb_004545_110 |
0.0750669876 |
- |
- |
DHHC-type zinc finger family protein isoform 1 [Theobroma cacao] |
| 11 |
Hb_003058_120 |
0.076940508 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 4 homolog [Jatropha curcas] |
| 12 |
Hb_000260_470 |
0.0773925189 |
- |
- |
PREDICTED: uncharacterized protein LOC105649044 [Jatropha curcas] |
| 13 |
Hb_164390_010 |
0.0774335839 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 14 |
Hb_001269_600 |
0.0775376628 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 15 |
Hb_007894_150 |
0.0775490479 |
- |
- |
hypothetical protein EUGRSUZ_F03462 [Eucalyptus grandis] |
| 16 |
Hb_000365_230 |
0.0783416673 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |
| 17 |
Hb_000853_150 |
0.0784500683 |
- |
- |
Fumarase 1 isoform 2 [Theobroma cacao] |
| 18 |
Hb_000167_010 |
0.0787258267 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_008616_050 |
0.0791725493 |
- |
- |
PREDICTED: putative deoxyribonuclease TATDN1 [Jatropha curcas] |
| 20 |
Hb_159809_070 |
0.0793285485 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |