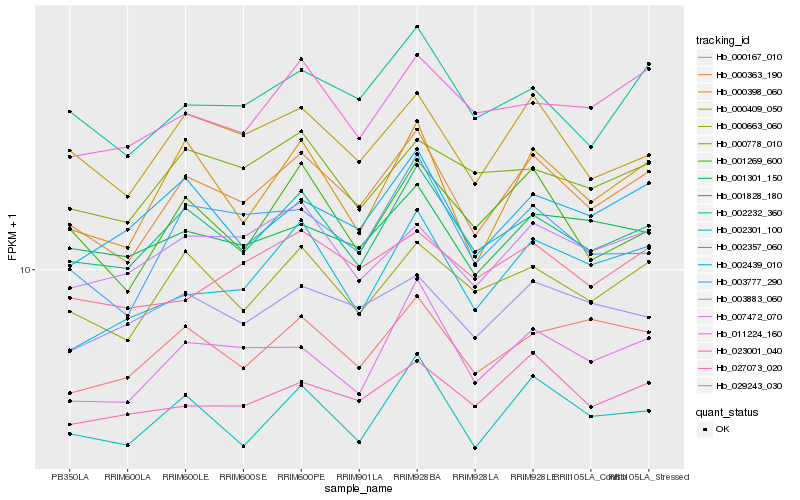
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000363_190 |
0.0 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-2-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_001828_180 |
0.0568666225 |
- |
- |
26S proteasome regulatory subunit S3, putative [Ricinus communis] |
| 3 |
Hb_000663_060 |
0.0630809686 |
- |
- |
hypothetical protein JCGZ_16277 [Jatropha curcas] |
| 4 |
Hb_000409_050 |
0.0641749674 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |
| 5 |
Hb_000398_060 |
0.068477347 |
- |
- |
PREDICTED: survival of motor neuron-related-splicing factor 30 [Jatropha curcas] |
| 6 |
Hb_002232_360 |
0.0696969276 |
- |
- |
PREDICTED: proline iminopeptidase isoform X2 [Jatropha curcas] |
| 7 |
Hb_002301_100 |
0.0699281981 |
- |
- |
hypothetical protein JCGZ_21479 [Jatropha curcas] |
| 8 |
Hb_003777_290 |
0.0731140946 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 9 |
Hb_011224_160 |
0.074306868 |
transcription factor, rubber biosynthesis |
TF Family: HSF, Gene Name: Farnesyl diphosphate synthase |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 10 |
Hb_001269_600 |
0.0767565097 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 11 |
Hb_027073_020 |
0.0778212561 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 12 |
Hb_007472_070 |
0.0791591969 |
- |
- |
cir, putative [Ricinus communis] |
| 13 |
Hb_000167_010 |
0.079434865 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002439_010 |
0.0799746497 |
- |
- |
PREDICTED: DNA damage-inducible protein 1 [Jatropha curcas] |
| 15 |
Hb_001301_150 |
0.0805500484 |
- |
- |
PREDICTED: UPF0136 membrane protein At2g26240 [Vitis vinifera] |
| 16 |
Hb_029243_030 |
0.0820835099 |
- |
- |
PREDICTED: regulatory-associated protein of TOR 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_023001_040 |
0.0823045973 |
- |
- |
Metallopeptidase M24 family protein isoform 1 [Theobroma cacao] |
| 18 |
Hb_000778_010 |
0.0834270024 |
- |
- |
hypothetical protein [Bacillus subtilis] |
| 19 |
Hb_002357_060 |
0.083483994 |
- |
- |
hypothetical protein CICLE_v10006112mg [Citrus clementina] |
| 20 |
Hb_003883_060 |
0.0839230519 |
transcription factor |
TF Family: BSD |
PREDICTED: probable RNA polymerase II transcription factor B subunit 1-1 isoform X1 [Jatropha curcas] |