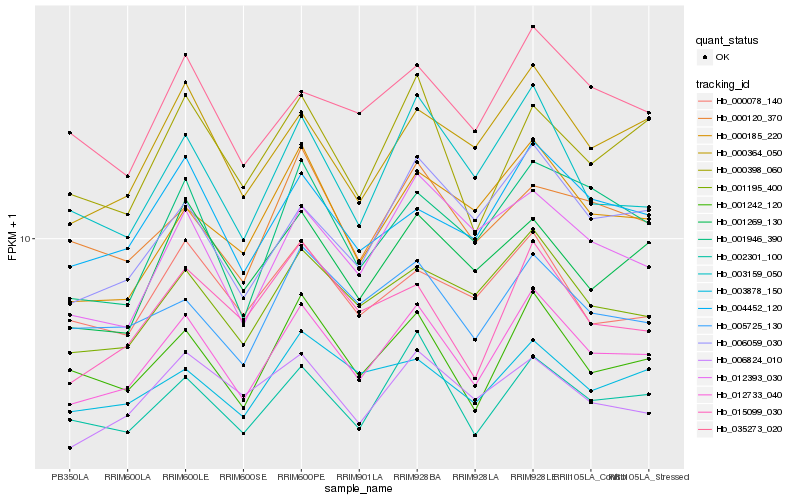
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012733_040 |
0.0 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 2 |
Hb_001242_120 |
0.0632302975 |
- |
- |
PREDICTED: carboxypeptidase D [Jatropha curcas] |
| 3 |
Hb_001195_400 |
0.0651724123 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 4 |
Hb_002301_100 |
0.0666875472 |
- |
- |
hypothetical protein JCGZ_21479 [Jatropha curcas] |
| 5 |
Hb_000364_050 |
0.0715463611 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_004452_120 |
0.0722097908 |
- |
- |
PREDICTED: uncharacterized protein LOC105639574 [Jatropha curcas] |
| 7 |
Hb_003159_050 |
0.0735073807 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 3, mitochondrial isoform X1 [Jatropha curcas] |
| 8 |
Hb_001946_390 |
0.0746944239 |
- |
- |
PREDICTED: NADH-cytochrome b5 reductase-like protein [Jatropha curcas] |
| 9 |
Hb_006059_030 |
0.0775307712 |
- |
- |
glutamate dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_000398_060 |
0.0788318711 |
- |
- |
PREDICTED: survival of motor neuron-related-splicing factor 30 [Jatropha curcas] |
| 11 |
Hb_005725_130 |
0.0792045966 |
- |
- |
PREDICTED: uncharacterized protein KIAA0930 homolog isoform X2 [Jatropha curcas] |
| 12 |
Hb_001269_130 |
0.079217055 |
- |
- |
plant poly(A)+ RNA export protein, putative [Ricinus communis] |
| 13 |
Hb_035273_020 |
0.080327309 |
- |
- |
PREDICTED: amidase 1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000185_220 |
0.0811483328 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 15 |
Hb_006824_010 |
0.0819101777 |
- |
- |
PREDICTED: origin of replication complex subunit 4 [Jatropha curcas] |
| 16 |
Hb_003878_150 |
0.0822706988 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 17 |
Hb_012393_030 |
0.0824689109 |
- |
- |
NADP-dependent isocitrate dehydrogenase family protein [Populus trichocarpa] |
| 18 |
Hb_015099_030 |
0.0830558825 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 19 |
Hb_000078_140 |
0.0843155848 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 20 |
Hb_000120_370 |
0.0845459519 |
- |
- |
type 2 diacylglycerol acyltransferase [Ricinus communis] |