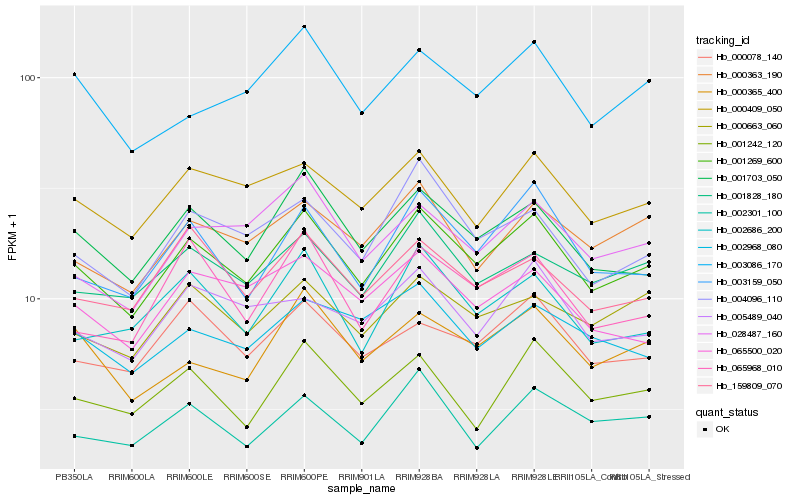
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001269_600 |
0.0 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 2 |
Hb_001703_050 |
0.0613659513 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 3 |
Hb_003159_050 |
0.0680474648 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 3, mitochondrial isoform X1 [Jatropha curcas] |
| 4 |
Hb_000409_050 |
0.0700641332 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |
| 5 |
Hb_001828_180 |
0.0736346653 |
- |
- |
26S proteasome regulatory subunit S3, putative [Ricinus communis] |
| 6 |
Hb_004096_110 |
0.073905074 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 7 |
Hb_000363_190 |
0.0767565097 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM22-2-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000078_140 |
0.0770643948 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 9 |
Hb_000663_060 |
0.0775376628 |
- |
- |
hypothetical protein JCGZ_16277 [Jatropha curcas] |
| 10 |
Hb_002686_200 |
0.0791833327 |
- |
- |
glucosidase II beta subunit, putative [Ricinus communis] |
| 11 |
Hb_000365_400 |
0.0798955244 |
- |
- |
metal ion transporter, putative [Ricinus communis] |
| 12 |
Hb_065500_020 |
0.080258344 |
- |
- |
Exocyst complex component sec3A isoform 1 [Theobroma cacao] |
| 13 |
Hb_028487_160 |
0.0807854114 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 14 |
Hb_159809_070 |
0.0809315597 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |
| 15 |
Hb_002301_100 |
0.0823998902 |
- |
- |
hypothetical protein JCGZ_21479 [Jatropha curcas] |
| 16 |
Hb_003086_170 |
0.0831086902 |
- |
- |
PREDICTED: putative phosphatidylglycerol/phosphatidylinositol transfer protein DDB_G0282179 [Jatropha curcas] |
| 17 |
Hb_001242_120 |
0.083292379 |
- |
- |
PREDICTED: carboxypeptidase D [Jatropha curcas] |
| 18 |
Hb_005489_040 |
0.0833471603 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |
| 19 |
Hb_065968_010 |
0.0837104561 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002968_080 |
0.0838356244 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 5 [Jatropha curcas] |