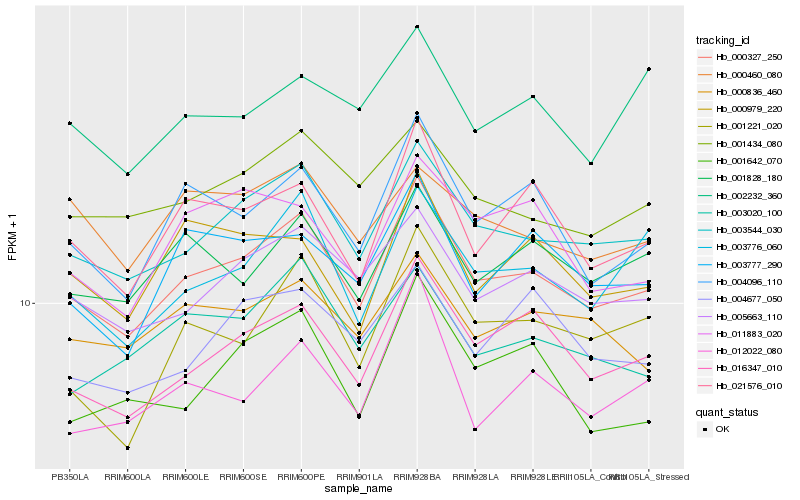
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000327_250 |
0.0 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 2 |
Hb_003544_030 |
0.0410077085 |
- |
- |
PREDICTED: citrate synthase, glyoxysomal [Jatropha curcas] |
| 3 |
Hb_016347_010 |
0.0604737353 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 4 |
Hb_005663_110 |
0.0619879082 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X2 [Jatropha curcas] |
| 5 |
Hb_004096_110 |
0.0660376411 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 6 |
Hb_021576_010 |
0.0690037984 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 7 |
Hb_001434_080 |
0.0721840831 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Jatropha curcas] |
| 8 |
Hb_003776_060 |
0.0745144948 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 9 |
Hb_001828_180 |
0.0765728859 |
- |
- |
26S proteasome regulatory subunit S3, putative [Ricinus communis] |
| 10 |
Hb_003777_290 |
0.0792956098 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 11 |
Hb_000836_460 |
0.0796099486 |
- |
- |
PREDICTED: uncharacterized protein LOC105628435 [Jatropha curcas] |
| 12 |
Hb_003020_100 |
0.0803271708 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 13 |
Hb_012022_080 |
0.0831739688 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 14 |
Hb_001221_020 |
0.0843393712 |
- |
- |
PREDICTED: probable aspartyl aminopeptidase [Jatropha curcas] |
| 15 |
Hb_001642_070 |
0.0845985353 |
- |
- |
PREDICTED: uncharacterized protein LOC105646431 [Jatropha curcas] |
| 16 |
Hb_000979_220 |
0.085348029 |
- |
- |
PREDICTED: uncharacterized protein LOC105634085 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004677_050 |
0.0862385861 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_011883_020 |
0.0865380885 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: aspartate--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 19 |
Hb_002232_360 |
0.0883669307 |
- |
- |
PREDICTED: proline iminopeptidase isoform X2 [Jatropha curcas] |
| 20 |
Hb_000460_080 |
0.0888598601 |
- |
- |
acyl-coenzyme A binding domain containing, putative [Ricinus communis] |