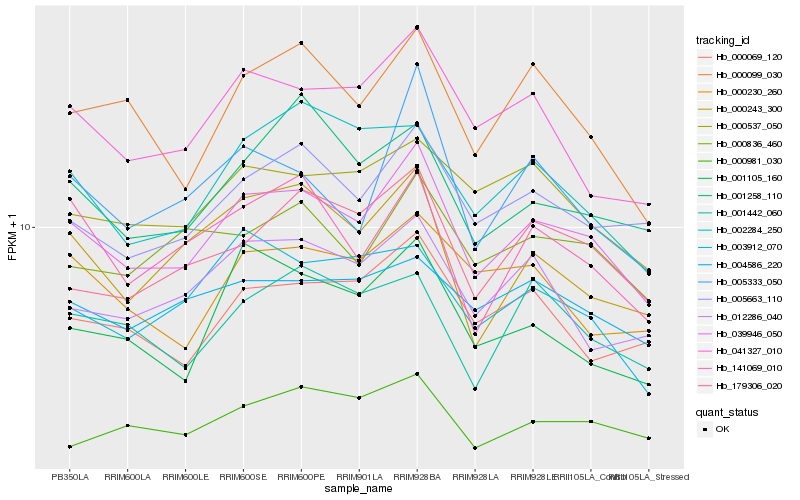
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_039946_050 |
0.0 |
- |
- |
catalytic, putative [Ricinus communis] |
| 2 |
Hb_005663_110 |
0.0681702119 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X2 [Jatropha curcas] |
| 3 |
Hb_041327_010 |
0.0735589172 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 4 |
Hb_179306_020 |
0.07569831 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000099_030 |
0.0767487634 |
- |
- |
6-phosphogluconate dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_000069_120 |
0.0779612927 |
- |
- |
PREDICTED: uncharacterized protein LOC105124742 isoform X1 [Populus euphratica] |
| 7 |
Hb_000537_050 |
0.0792855424 |
- |
- |
AP-2 complex subunit beta-1, putative [Ricinus communis] |
| 8 |
Hb_000836_460 |
0.0794009344 |
- |
- |
PREDICTED: uncharacterized protein LOC105628435 [Jatropha curcas] |
| 9 |
Hb_141069_010 |
0.0797885306 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_012286_040 |
0.0798781694 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 11 |
Hb_000243_300 |
0.0816639172 |
- |
- |
catalytic, putative [Ricinus communis] |
| 12 |
Hb_001105_160 |
0.0817589903 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 13 |
Hb_004586_220 |
0.081827715 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 14 |
Hb_000981_030 |
0.0820989567 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105640649 [Jatropha curcas] |
| 15 |
Hb_001442_060 |
0.0830686708 |
- |
- |
PREDICTED: uncharacterized protein LOC105638091 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001258_110 |
0.0838421809 |
- |
- |
transporter, putative [Ricinus communis] |
| 17 |
Hb_005333_050 |
0.0843171153 |
- |
- |
PREDICTED: glucose-6-phosphate isomerase 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000230_260 |
0.0845934466 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 19 |
Hb_002284_250 |
0.0846128253 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 20 |
Hb_003912_070 |
0.085173804 |
- |
- |
PREDICTED: nuclear pore complex protein NUP85 [Jatropha curcas] |