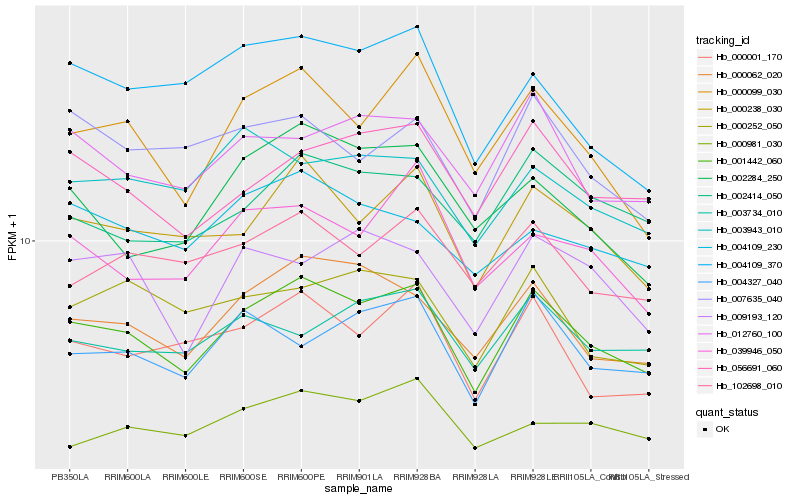
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001442_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638091 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000062_020 |
0.0607841401 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 3 |
Hb_000238_030 |
0.0628460332 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_000099_030 |
0.0688187181 |
- |
- |
6-phosphogluconate dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_056691_060 |
0.0738256588 |
- |
- |
PREDICTED: probable manganese-transporting ATPase PDR2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000001_170 |
0.0738516061 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 7 |
Hb_012760_100 |
0.0753964001 |
- |
- |
Pre-rRNA-processing protein ESF1, putative [Ricinus communis] |
| 8 |
Hb_002414_050 |
0.0773629678 |
- |
- |
hypothetical protein CICLE_v10000243mg [Citrus clementina] |
| 9 |
Hb_102698_010 |
0.0785826294 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_004327_040 |
0.0797699902 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 27 homolog [Jatropha curcas] |
| 11 |
Hb_039946_050 |
0.0830686708 |
- |
- |
catalytic, putative [Ricinus communis] |
| 12 |
Hb_004109_230 |
0.0831493383 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_009193_120 |
0.0833739967 |
desease resistance |
Gene Name: AAA_22 |
PREDICTED: putative ATP-dependent RNA helicase DHX33 [Jatropha curcas] |
| 14 |
Hb_002284_250 |
0.0834422575 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 15 |
Hb_000252_050 |
0.0834666646 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |
| 16 |
Hb_007635_040 |
0.084808542 |
- |
- |
PREDICTED: WAT1-related protein At3g45870 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004109_370 |
0.0853979516 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 18 |
Hb_003943_010 |
0.0865766081 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000981_030 |
0.0868877778 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105640649 [Jatropha curcas] |
| 20 |
Hb_003734_010 |
0.0870113398 |
- |
- |
PREDICTED: double-strand break repair protein MRE11 [Jatropha curcas] |