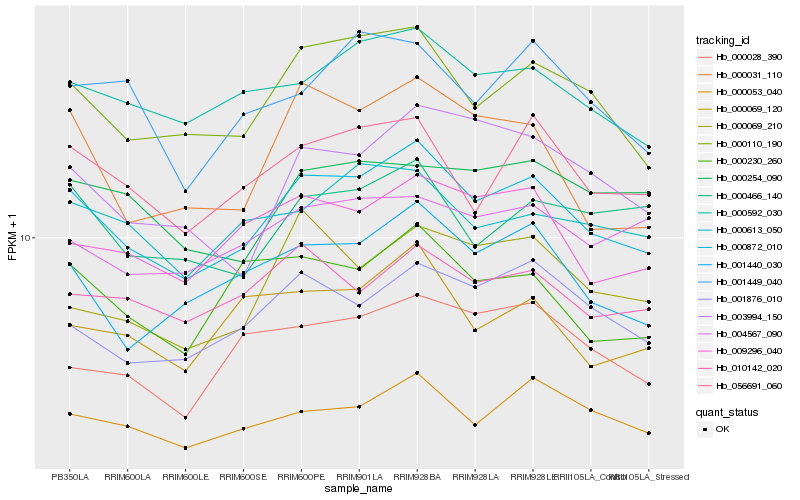
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000872_010 |
0.0 |
- |
- |
PREDICTED: putative ubiquitin-conjugating enzyme E2 38 [Jatropha curcas] |
| 2 |
Hb_000110_190 |
0.0560747982 |
- |
- |
hypothetical protein B456_013G081300 [Gossypium raimondii] |
| 3 |
Hb_001440_030 |
0.0665685765 |
- |
- |
PREDICTED: exocyst complex component SEC8 [Jatropha curcas] |
| 4 |
Hb_000592_030 |
0.0668355019 |
- |
- |
PREDICTED: uncharacterized protein LOC100853969 isoform X1 [Vitis vinifera] |
| 5 |
Hb_056691_060 |
0.0682526364 |
- |
- |
PREDICTED: probable manganese-transporting ATPase PDR2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000053_040 |
0.0711551345 |
- |
- |
hypothetical protein CISIN_1g0010392mg, partial [Citrus sinensis] |
| 7 |
Hb_001876_010 |
0.0720581186 |
- |
- |
PREDICTED: arginyl-tRNA--protein transferase 2-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_009296_040 |
0.0729766223 |
- |
- |
hypothetical protein L484_007435 [Morus notabilis] |
| 9 |
Hb_000230_260 |
0.0778390644 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 10 |
Hb_010142_020 |
0.0788118182 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003994_150 |
0.0796120688 |
- |
- |
PREDICTED: pyrophosphate-energized membrane proton pump 2 [Jatropha curcas] |
| 12 |
Hb_000031_110 |
0.0799546794 |
- |
- |
Coatomer, beta subunit isoform 1 [Theobroma cacao] |
| 13 |
Hb_000613_050 |
0.0800283046 |
- |
- |
PREDICTED: uncharacterized protein LOC105641544 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000069_120 |
0.0813743377 |
- |
- |
PREDICTED: uncharacterized protein LOC105124742 isoform X1 [Populus euphratica] |
| 15 |
Hb_000466_140 |
0.0814272283 |
- |
- |
hypothetical protein JCGZ_02122 [Jatropha curcas] |
| 16 |
Hb_000028_390 |
0.0819387404 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 3-like [Jatropha curcas] |
| 17 |
Hb_004567_090 |
0.0830230738 |
- |
- |
PREDICTED: GPI transamidase component PIG-T [Jatropha curcas] |
| 18 |
Hb_000069_210 |
0.0835143828 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 19 |
Hb_001449_040 |
0.084058788 |
- |
- |
PREDICTED: SWR1 complex subunit 2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000254_090 |
0.0843416862 |
- |
- |
PREDICTED: la protein 2 [Jatropha curcas] |