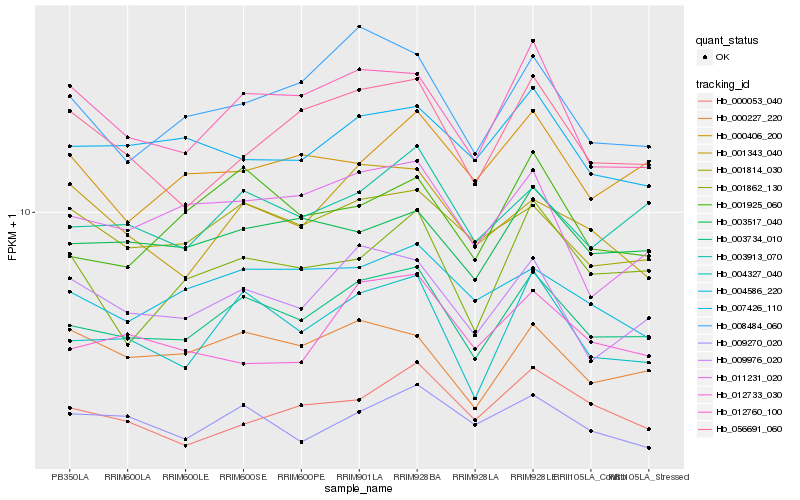
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003734_010 |
0.0 |
- |
- |
PREDICTED: double-strand break repair protein MRE11 [Jatropha curcas] |
| 2 |
Hb_004327_040 |
0.0468586292 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 27 homolog [Jatropha curcas] |
| 3 |
Hb_003913_070 |
0.0512291645 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 30 [Populus euphratica] |
| 4 |
Hb_001814_030 |
0.0566257913 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 5 |
Hb_009976_020 |
0.0567953495 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105631286 [Jatropha curcas] |
| 6 |
Hb_003517_040 |
0.0576722427 |
- |
- |
PREDICTED: putative DUF21 domain-containing protein At3g13070, chloroplastic [Jatropha curcas] |
| 7 |
Hb_012760_100 |
0.0595759808 |
- |
- |
Pre-rRNA-processing protein ESF1, putative [Ricinus communis] |
| 8 |
Hb_008484_060 |
0.0619060754 |
- |
- |
PREDICTED: allantoate deiminase isoform X3 [Populus euphratica] |
| 9 |
Hb_004586_220 |
0.0633960204 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 10 |
Hb_000227_220 |
0.0645930882 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 11 |
Hb_007426_110 |
0.0650542853 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X2 [Jatropha curcas] |
| 12 |
Hb_009270_020 |
0.0654781084 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: helicase and polymerase-containing protein TEBICHI [Jatropha curcas] |
| 13 |
Hb_011231_020 |
0.0659699413 |
- |
- |
PREDICTED: proline-, glutamic acid- and leucine-rich protein 1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001925_060 |
0.066448464 |
- |
- |
PREDICTED: putative RNA polymerase II subunit B1 CTD phosphatase RPAP2 homolog [Jatropha curcas] |
| 15 |
Hb_056691_060 |
0.0677708166 |
- |
- |
PREDICTED: probable manganese-transporting ATPase PDR2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000053_040 |
0.0697162079 |
- |
- |
hypothetical protein CISIN_1g0010392mg, partial [Citrus sinensis] |
| 17 |
Hb_000406_200 |
0.0706481479 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit epsilon-like isoform X4 [Jatropha curcas] |
| 18 |
Hb_001862_130 |
0.0721518329 |
- |
- |
Protein FRIGIDA, putative [Ricinus communis] |
| 19 |
Hb_001343_040 |
0.0724550785 |
- |
- |
PREDICTED: uncharacterized protein LOC105638555 [Jatropha curcas] |
| 20 |
Hb_012733_030 |
0.072476388 |
- |
- |
hypothetical protein OsI_15243 [Oryza sativa Indica Group] |