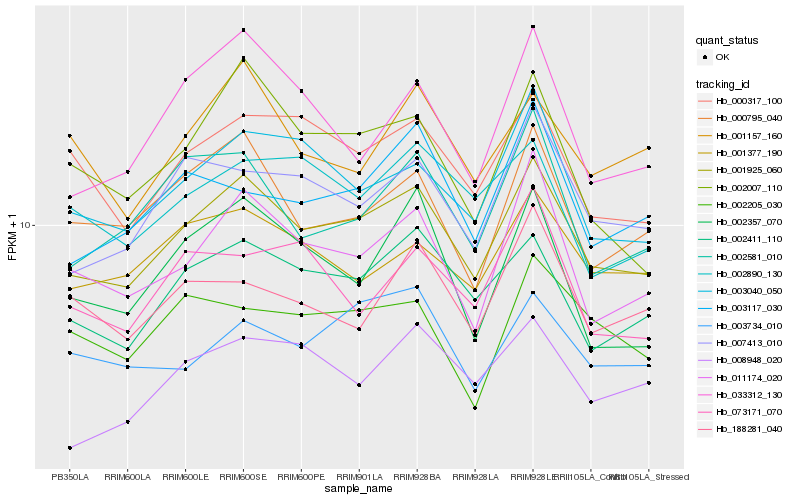
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001925_060 |
0.0 |
- |
- |
PREDICTED: putative RNA polymerase II subunit B1 CTD phosphatase RPAP2 homolog [Jatropha curcas] |
| 2 |
Hb_002411_110 |
0.0486791108 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC2 [Jatropha curcas] |
| 3 |
Hb_003040_050 |
0.0565103231 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 4 |
Hb_007413_010 |
0.0575347645 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 5 |
Hb_011174_020 |
0.0575762318 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 6 |
Hb_000317_100 |
0.0611713742 |
- |
- |
PREDICTED: uncharacterized protein LOC105646995 [Jatropha curcas] |
| 7 |
Hb_000795_040 |
0.0630811266 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 8 |
Hb_001377_190 |
0.0633283562 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 9 |
Hb_008948_020 |
0.0646031332 |
- |
- |
hypothetical protein JCGZ_21216 [Jatropha curcas] |
| 10 |
Hb_003734_010 |
0.066448464 |
- |
- |
PREDICTED: double-strand break repair protein MRE11 [Jatropha curcas] |
| 11 |
Hb_002581_010 |
0.0668281069 |
- |
- |
PREDICTED: uncharacterized protein LOC105641469 isoform X2 [Jatropha curcas] |
| 12 |
Hb_003117_030 |
0.0672939653 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 13 |
Hb_002890_130 |
0.0703326836 |
- |
- |
tip120, putative [Ricinus communis] |
| 14 |
Hb_033312_130 |
0.0707626895 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001157_160 |
0.0714523078 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BRE1-like 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002205_030 |
0.0720846345 |
- |
- |
PREDICTED: alpha-galactosidase 3 [Jatropha curcas] |
| 17 |
Hb_002357_070 |
0.0721205392 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 18 |
Hb_002007_110 |
0.0723222499 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 19 |
Hb_073171_070 |
0.0735129679 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 20 |
Hb_188281_040 |
0.0735319853 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12 [Jatropha curcas] |