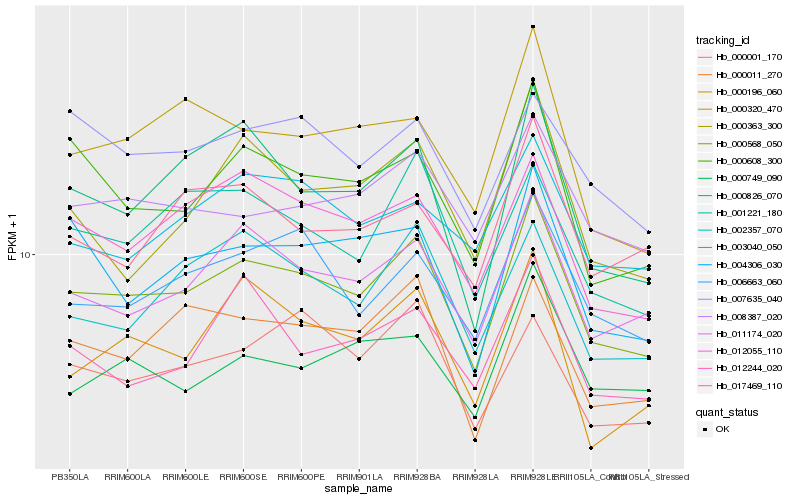
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000568_050 |
0.0 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_011174_020 |
0.0563978432 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 3 |
Hb_012055_110 |
0.0667851602 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 4 |
Hb_006663_060 |
0.0675755118 |
- |
- |
PREDICTED: calcium homeostasis endoplasmic reticulum protein [Jatropha curcas] |
| 5 |
Hb_000011_270 |
0.0676342982 |
- |
- |
PREDICTED: pumilio homolog 23 [Jatropha curcas] |
| 6 |
Hb_000608_300 |
0.0691054539 |
- |
- |
PREDICTED: protein KRI1 homolog [Jatropha curcas] |
| 7 |
Hb_000826_070 |
0.0705306288 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_008387_020 |
0.0711596069 |
- |
- |
PREDICTED: FHA domain-containing protein DDL isoform X1 [Jatropha curcas] |
| 9 |
Hb_001221_180 |
0.0714676475 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 10 |
Hb_002357_070 |
0.0729589396 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 11 |
Hb_004306_030 |
0.0747419178 |
- |
- |
PREDICTED: GPI transamidase component PIG-S [Jatropha curcas] |
| 12 |
Hb_000320_470 |
0.0760029149 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 13 |
Hb_000196_060 |
0.0776926104 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 14 |
Hb_007635_040 |
0.077693772 |
- |
- |
PREDICTED: WAT1-related protein At3g45870 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000001_170 |
0.0783136149 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 16 |
Hb_000749_090 |
0.0787769273 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_003040_050 |
0.078828346 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 18 |
Hb_012244_020 |
0.0788754647 |
- |
- |
calpain, putative [Ricinus communis] |
| 19 |
Hb_017469_110 |
0.0799272437 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 20 |
Hb_000363_300 |
0.0803541446 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |