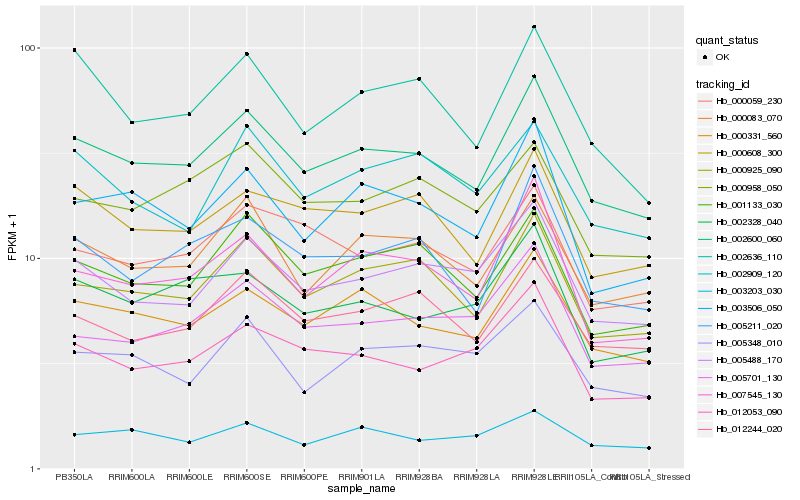
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002600_060 |
0.0 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 2 |
Hb_000331_560 |
0.0529647632 |
transcription factor |
TF Family: C2H2 |
PREDICTED: lysine-specific demethylase REF6 [Jatropha curcas] |
| 3 |
Hb_000925_090 |
0.0549595632 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005211_020 |
0.0667850467 |
- |
- |
PREDICTED: serine/threonine-protein kinase prpf4B-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001133_030 |
0.0685846842 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 6 |
Hb_005488_170 |
0.0687262627 |
- |
- |
PREDICTED: septin and tuftelin-interacting protein 1 homolog 1 [Jatropha curcas] |
| 7 |
Hb_000083_070 |
0.0693664863 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_012244_020 |
0.0700983276 |
- |
- |
calpain, putative [Ricinus communis] |
| 9 |
Hb_000608_300 |
0.0707505435 |
- |
- |
PREDICTED: protein KRI1 homolog [Jatropha curcas] |
| 10 |
Hb_007545_130 |
0.0737749784 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_003506_050 |
0.074622406 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_002636_110 |
0.0771329692 |
- |
- |
PREDICTED: heat shock protein 83-like isoform X2 [Gossypium raimondii] |
| 13 |
Hb_000059_230 |
0.0795885714 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |
| 14 |
Hb_012053_090 |
0.0812707222 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105637346 [Jatropha curcas] |
| 15 |
Hb_002328_040 |
0.0820581084 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g53700, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000958_050 |
0.0823775767 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 17 |
Hb_005701_130 |
0.0825154085 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |
| 18 |
Hb_005348_010 |
0.0837936187 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003203_030 |
0.086404352 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g13600-like [Jatropha curcas] |
| 20 |
Hb_002909_120 |
0.0864833269 |
- |
- |
PREDICTED: transcription elongation factor SPT6 [Jatropha curcas] |