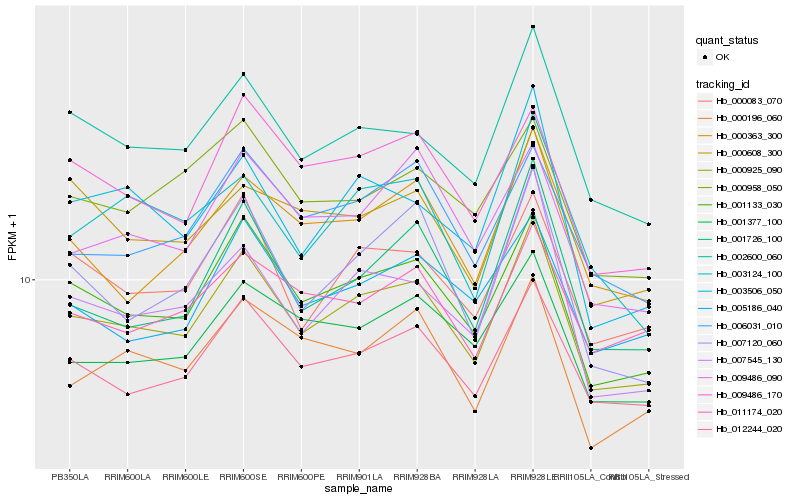
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000925_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001133_030 |
0.0385031694 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 3 |
Hb_012244_020 |
0.0518010489 |
- |
- |
calpain, putative [Ricinus communis] |
| 4 |
Hb_002600_060 |
0.0549595632 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 5 |
Hb_000083_070 |
0.0608588003 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_009486_170 |
0.0685842474 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 7 |
Hb_001726_100 |
0.0694748473 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 19 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001377_100 |
0.0702738306 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000363_300 |
0.0707816127 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_006031_010 |
0.0720084712 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X3 [Jatropha curcas] |
| 11 |
Hb_007120_060 |
0.0720552576 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 12 |
Hb_000196_060 |
0.072326729 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 13 |
Hb_007545_130 |
0.0727864195 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_005186_040 |
0.0737626392 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 15 |
Hb_003506_050 |
0.0751977899 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_011174_020 |
0.0755921327 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 17 |
Hb_009486_090 |
0.0760972125 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_003124_100 |
0.0761888725 |
- |
- |
- |
| 19 |
Hb_000608_300 |
0.0768319934 |
- |
- |
PREDICTED: protein KRI1 homolog [Jatropha curcas] |
| 20 |
Hb_000958_050 |
0.077715595 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |