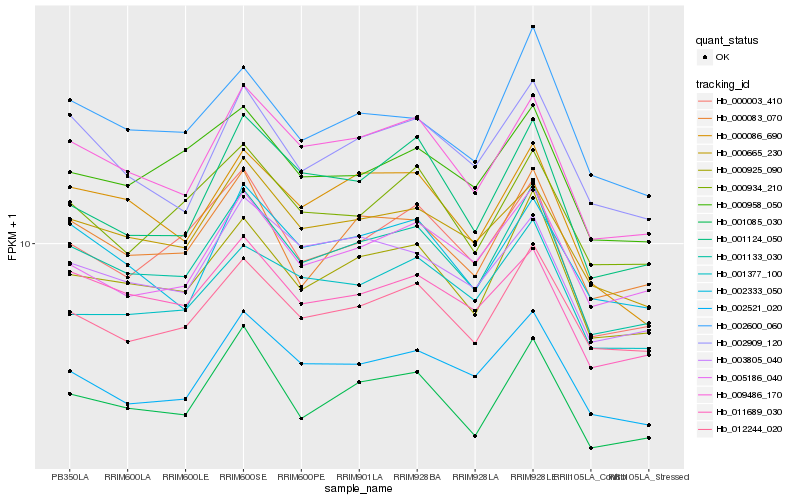
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001133_030 |
0.0 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 2 |
Hb_000925_090 |
0.0385031694 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 3 |
Hb_009486_170 |
0.0389470421 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 4 |
Hb_012244_020 |
0.0542870003 |
- |
- |
calpain, putative [Ricinus communis] |
| 5 |
Hb_011689_030 |
0.0592509887 |
- |
- |
80 kD MCM3-associated protein, putative [Ricinus communis] |
| 6 |
Hb_000003_410 |
0.0601394183 |
- |
- |
PREDICTED: intron-binding protein aquarius [Jatropha curcas] |
| 7 |
Hb_002521_020 |
0.0603663489 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03100, mitochondrial-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_000665_230 |
0.0606210826 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 9 |
Hb_000083_070 |
0.0606356602 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_003805_040 |
0.0612976574 |
- |
- |
Sacsin [Glycine soja] |
| 11 |
Hb_000086_690 |
0.0659930112 |
- |
- |
PREDICTED: splicing factor 3B subunit 1 [Jatropha curcas] |
| 12 |
Hb_005186_040 |
0.0671615985 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 13 |
Hb_002909_120 |
0.0671906842 |
- |
- |
PREDICTED: transcription elongation factor SPT6 [Jatropha curcas] |
| 14 |
Hb_001124_050 |
0.0672807185 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 15 |
Hb_000958_050 |
0.0680551209 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 16 |
Hb_002333_050 |
0.0684688367 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002600_060 |
0.0685846842 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 18 |
Hb_000934_210 |
0.0693100193 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 19 |
Hb_001377_100 |
0.0705164388 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001085_030 |
0.0710606977 |
- |
- |
Tetratricopeptide repeat-like superfamily protein, putative isoform 1 [Theobroma cacao] |