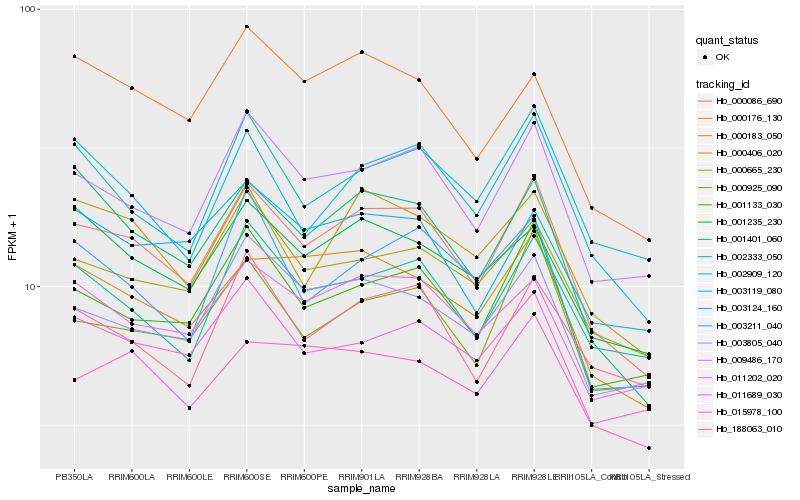
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000086_690 |
0.0 |
- |
- |
PREDICTED: splicing factor 3B subunit 1 [Jatropha curcas] |
| 2 |
Hb_009486_170 |
0.0569590964 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 3 |
Hb_001133_030 |
0.0659930112 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 4 |
Hb_000183_050 |
0.0713080373 |
- |
- |
PREDICTED: alpha-mannosidase 2 [Jatropha curcas] |
| 5 |
Hb_003119_080 |
0.0717220669 |
- |
- |
PREDICTED: splicing factor 3B subunit 1 [Jatropha curcas] |
| 6 |
Hb_001401_060 |
0.0727442652 |
- |
- |
PREDICTED: protein WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 1-like [Jatropha curcas] |
| 7 |
Hb_011202_020 |
0.0737011522 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000406_020 |
0.0738999333 |
- |
- |
- |
| 9 |
Hb_000665_230 |
0.0739920815 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 10 |
Hb_003805_040 |
0.0754773647 |
- |
- |
Sacsin [Glycine soja] |
| 11 |
Hb_002333_050 |
0.0767993952 |
- |
- |
PREDICTED: uncharacterized protein LOC105644854 isoform X1 [Jatropha curcas] |
| 12 |
Hb_011689_030 |
0.0771495184 |
- |
- |
80 kD MCM3-associated protein, putative [Ricinus communis] |
| 13 |
Hb_000925_090 |
0.0783187922 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001235_230 |
0.0787089367 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 15 |
Hb_000176_130 |
0.0787560777 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Jatropha curcas] |
| 16 |
Hb_015978_100 |
0.0827520468 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH6 [Jatropha curcas] |
| 17 |
Hb_188063_010 |
0.0830738715 |
- |
- |
hypothetical protein RCOM_1469330 [Ricinus communis] |
| 18 |
Hb_003124_160 |
0.0834130816 |
- |
- |
dynamin, putative [Ricinus communis] |
| 19 |
Hb_003211_040 |
0.0841512101 |
- |
- |
PREDICTED: uncharacterized protein LOC105634664 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002909_120 |
0.0843739774 |
- |
- |
PREDICTED: transcription elongation factor SPT6 [Jatropha curcas] |