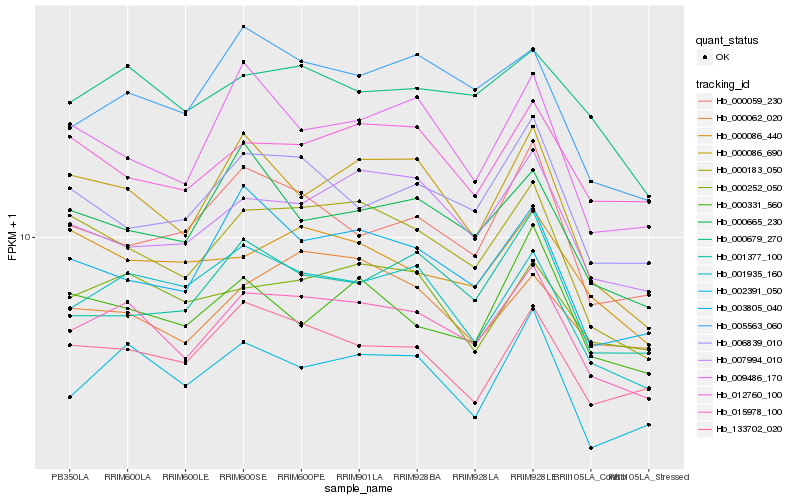
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015978_100 |
0.0 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH6 [Jatropha curcas] |
| 2 |
Hb_000183_050 |
0.0681881927 |
- |
- |
PREDICTED: alpha-mannosidase 2 [Jatropha curcas] |
| 3 |
Hb_000679_270 |
0.0727183442 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 4 |
Hb_001935_160 |
0.0784613476 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 5 |
Hb_002391_050 |
0.080039641 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17670 [Jatropha curcas] |
| 6 |
Hb_005563_060 |
0.0823800995 |
- |
- |
PREDICTED: coatomer subunit alpha-1 [Jatropha curcas] |
| 7 |
Hb_000252_050 |
0.082544581 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |
| 8 |
Hb_000086_690 |
0.0827520468 |
- |
- |
PREDICTED: splicing factor 3B subunit 1 [Jatropha curcas] |
| 9 |
Hb_133702_020 |
0.0838607137 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_007994_010 |
0.084154199 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003805_040 |
0.0873231147 |
- |
- |
Sacsin [Glycine soja] |
| 12 |
Hb_006839_010 |
0.0875259765 |
- |
- |
PREDICTED: importin subunit beta-1-like [Jatropha curcas] |
| 13 |
Hb_001377_100 |
0.0904253887 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000086_440 |
0.0906152868 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 15 |
Hb_000059_230 |
0.0906303845 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |
| 16 |
Hb_012760_100 |
0.090765267 |
- |
- |
Pre-rRNA-processing protein ESF1, putative [Ricinus communis] |
| 17 |
Hb_000062_020 |
0.0925767786 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 18 |
Hb_000665_230 |
0.0927660504 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 19 |
Hb_009486_170 |
0.0944089654 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 20 |
Hb_000331_560 |
0.0947177291 |
transcription factor |
TF Family: C2H2 |
PREDICTED: lysine-specific demethylase REF6 [Jatropha curcas] |