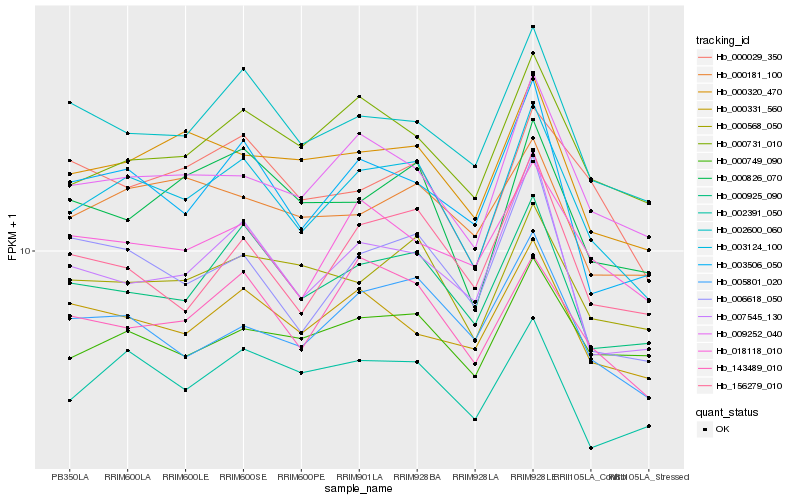
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003124_100 |
0.0 |
- |
- |
- |
| 2 |
Hb_000925_090 |
0.0761888725 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 3 |
Hb_009252_040 |
0.0821791318 |
- |
- |
PREDICTED: protein EXECUTER 2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_143489_010 |
0.0863425643 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 50 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000320_470 |
0.0883967363 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 6 |
Hb_002600_060 |
0.0884191097 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 7 |
Hb_003506_050 |
0.0884793301 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_000731_010 |
0.0887296371 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000568_050 |
0.091755006 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_005801_020 |
0.0922738971 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 [Jatropha curcas] |
| 11 |
Hb_002391_050 |
0.0924047364 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17670 [Jatropha curcas] |
| 12 |
Hb_000826_070 |
0.0946643603 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000331_560 |
0.0953619844 |
transcription factor |
TF Family: C2H2 |
PREDICTED: lysine-specific demethylase REF6 [Jatropha curcas] |
| 14 |
Hb_007545_130 |
0.0958411382 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_018118_010 |
0.0978253642 |
- |
- |
DNA polymerase I, putative [Ricinus communis] |
| 16 |
Hb_000029_350 |
0.0979165735 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
4-hydroxy-3-methylbut-2-enyl diphosphate reductase [Hevea brasiliensis] |
| 17 |
Hb_000749_090 |
0.1000471913 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_006618_050 |
0.100794219 |
- |
- |
PREDICTED: elongation factor G-2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_156279_010 |
0.1008874909 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105641053 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000181_100 |
0.1015717168 |
- |
- |
PREDICTED: protein starmaker [Jatropha curcas] |