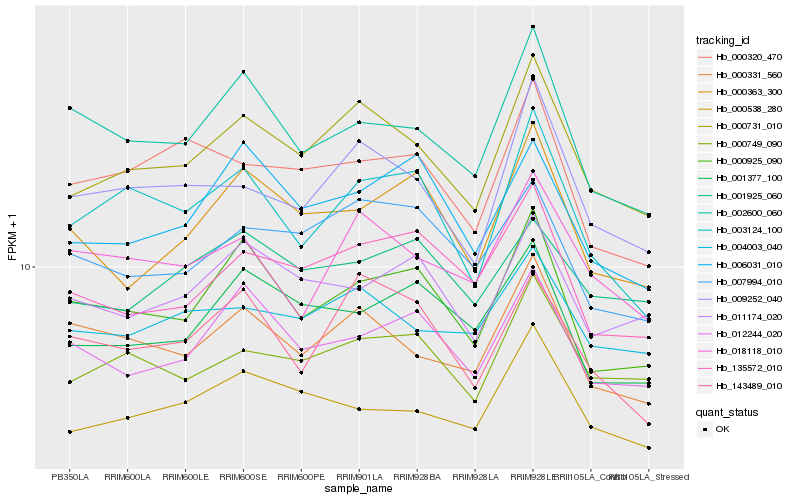
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000731_010 |
0.0 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_009252_040 |
0.0684321653 |
- |
- |
PREDICTED: protein EXECUTER 2, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000749_090 |
0.0713197288 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_004003_040 |
0.0765337545 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 5 |
Hb_000538_280 |
0.0795486013 |
- |
- |
PREDICTED: uncharacterized protein LOC105642112 [Jatropha curcas] |
| 6 |
Hb_000331_560 |
0.0822253774 |
transcription factor |
TF Family: C2H2 |
PREDICTED: lysine-specific demethylase REF6 [Jatropha curcas] |
| 7 |
Hb_135572_010 |
0.085962145 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 8 |
Hb_007994_010 |
0.0871505865 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000925_090 |
0.0876086668 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003124_100 |
0.0887296371 |
- |
- |
- |
| 11 |
Hb_018118_010 |
0.090035349 |
- |
- |
DNA polymerase I, putative [Ricinus communis] |
| 12 |
Hb_011174_020 |
0.0927837686 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 13 |
Hb_143489_010 |
0.0930465966 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 50 isoform X2 [Jatropha curcas] |
| 14 |
Hb_012244_020 |
0.093517095 |
- |
- |
calpain, putative [Ricinus communis] |
| 15 |
Hb_001377_100 |
0.0960890656 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000363_300 |
0.0963148038 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002600_060 |
0.0969489676 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 18 |
Hb_000320_470 |
0.0971501635 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 19 |
Hb_006031_010 |
0.0976482209 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X3 [Jatropha curcas] |
| 20 |
Hb_001925_060 |
0.0977384652 |
- |
- |
PREDICTED: putative RNA polymerase II subunit B1 CTD phosphatase RPAP2 homolog [Jatropha curcas] |