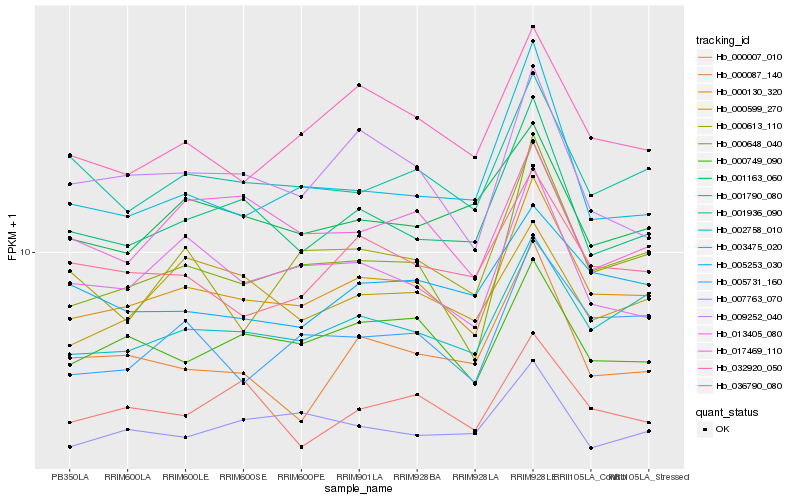
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000130_320 |
0.0 |
desease resistance |
Gene Name: AAA |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 2 |
Hb_000648_040 |
0.0631250469 |
- |
- |
unknown [Populus trichocarpa] |
| 3 |
Hb_002758_010 |
0.0668992322 |
- |
- |
PREDICTED: ribonuclease II, chloroplastic/mitochondrial [Populus euphratica] |
| 4 |
Hb_001790_080 |
0.0699293693 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic isoform X3 [Jatropha curcas] |
| 5 |
Hb_032920_050 |
0.0755077329 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000749_090 |
0.0763064662 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_001936_090 |
0.0789644198 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_005731_160 |
0.0792488711 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_013405_080 |
0.0815243024 |
- |
- |
PREDICTED: proline--tRNA ligase [Jatropha curcas] |
| 10 |
Hb_036790_080 |
0.0827098883 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 39 [Jatropha curcas] |
| 11 |
Hb_003475_020 |
0.0833939203 |
- |
- |
hypothetical protein JCGZ_13177 [Jatropha curcas] |
| 12 |
Hb_009252_040 |
0.0860105291 |
- |
- |
PREDICTED: protein EXECUTER 2, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000613_110 |
0.0890235263 |
- |
- |
PREDICTED: uncharacterized protein LOC105641540 [Jatropha curcas] |
| 14 |
Hb_007763_070 |
0.0919709228 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71460, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000007_010 |
0.0923798873 |
- |
- |
PREDICTED: protein PHYLLO, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001163_060 |
0.0923905776 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 17 |
Hb_000087_140 |
0.0940936918 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27270 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005253_030 |
0.0941620523 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_017469_110 |
0.0970157798 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 20 |
Hb_000599_270 |
0.0982463487 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A regulatory subunit B''beta isoform X3 [Jatropha curcas] |