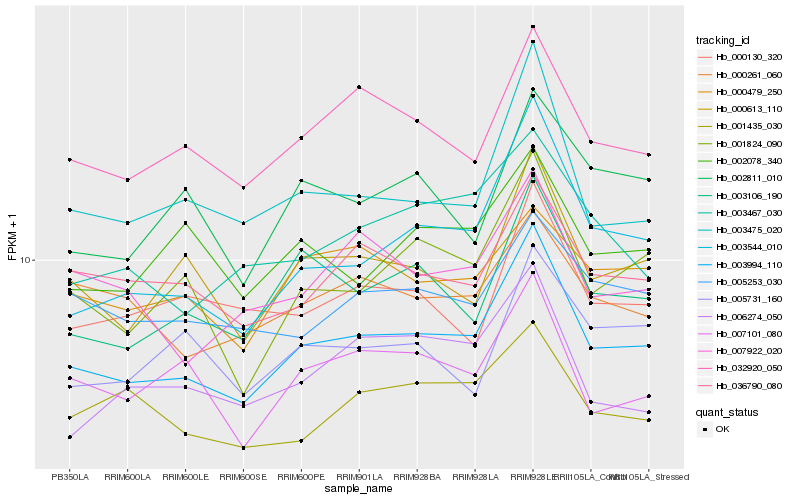
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003994_110 |
0.0 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646130 [Jatropha curcas] |
| 2 |
Hb_003544_010 |
0.0760515693 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit A, chloroplastic/mitochondrial [Jatropha curcas] |
| 3 |
Hb_003475_020 |
0.0816521116 |
- |
- |
hypothetical protein JCGZ_13177 [Jatropha curcas] |
| 4 |
Hb_000613_110 |
0.0826839088 |
- |
- |
PREDICTED: uncharacterized protein LOC105641540 [Jatropha curcas] |
| 5 |
Hb_032920_050 |
0.0832879228 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001824_090 |
0.0835064126 |
- |
- |
- |
| 7 |
Hb_003106_190 |
0.0841416574 |
- |
- |
Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA, putative [Ricinus communis] |
| 8 |
Hb_036790_080 |
0.0922669351 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 39 [Jatropha curcas] |
| 9 |
Hb_007101_080 |
0.096835074 |
transcription factor |
TF Family: GNAT |
GCN5-related N-acetyltransferase family protein [Populus trichocarpa] |
| 10 |
Hb_007922_020 |
0.0979257944 |
- |
- |
PREDICTED: uncharacterized protein LOC105646505 [Jatropha curcas] |
| 11 |
Hb_003467_030 |
0.1006711776 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 12 |
Hb_000130_320 |
0.1013329092 |
desease resistance |
Gene Name: AAA |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 13 |
Hb_005253_030 |
0.1020580272 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_002078_340 |
0.1036813675 |
- |
- |
PREDICTED: uncharacterized protein LOC105644820 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000261_060 |
0.1037308402 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Jatropha curcas] |
| 16 |
Hb_002811_010 |
0.1059943189 |
- |
- |
cysteine synthase, putative [Ricinus communis] |
| 17 |
Hb_001435_030 |
0.1070434982 |
- |
- |
PREDICTED: uncharacterized protein LOC105645073 [Jatropha curcas] |
| 18 |
Hb_000479_250 |
0.1079694235 |
- |
- |
PREDICTED: uncharacterized protein LOC105644121 [Jatropha curcas] |
| 19 |
Hb_006274_050 |
0.1093416398 |
- |
- |
hypothetical protein VITISV_022618 [Vitis vinifera] |
| 20 |
Hb_005731_160 |
0.1100663495 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |