| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
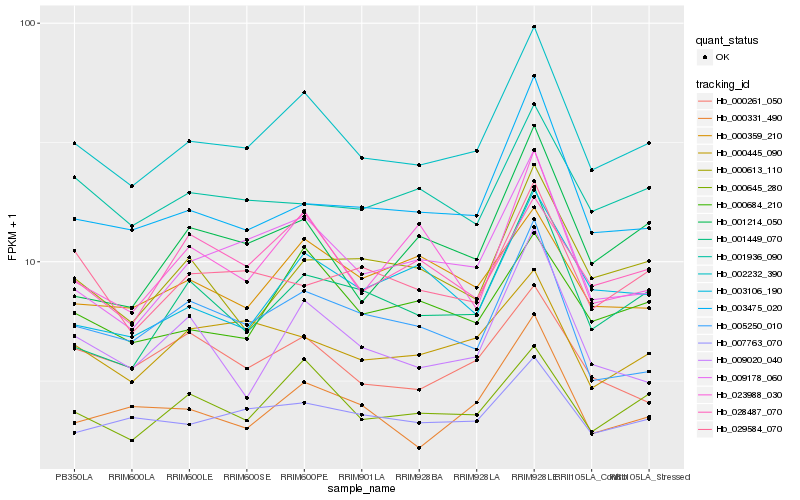
Hb_002232_390 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635994 [Jatropha curcas] |
| 2 |
Hb_009178_060 |
0.0744423811 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At4g31390, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_000645_280 |
0.0807691525 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 4 |
Hb_003475_020 |
0.0877375562 |
- |
- |
hypothetical protein JCGZ_13177 [Jatropha curcas] |
| 5 |
Hb_000684_210 |
0.0904512309 |
- |
- |
PREDICTED: uncharacterized protein LOC105642177 [Jatropha curcas] |
| 6 |
Hb_007763_070 |
0.0911377938 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71460, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001449_070 |
0.0925080639 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000331_490 |
0.0971448371 |
- |
- |
PREDICTED: radical S-adenosyl methionine domain-containing protein 1, mitochondrial [Jatropha curcas] |
| 9 |
Hb_005250_010 |
0.0973892255 |
- |
- |
hypothetical protein JCGZ_15339 [Jatropha curcas] |
| 10 |
Hb_000445_090 |
0.0984134432 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 11 |
Hb_003106_190 |
0.0998185547 |
- |
- |
Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA, putative [Ricinus communis] |
| 12 |
Hb_001214_050 |
0.1005783555 |
- |
- |
PREDICTED: bifunctional monothiol glutaredoxin-S16, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001936_090 |
0.1016805041 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_029584_070 |
0.1018068225 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15690-like [Jatropha curcas] |
| 15 |
Hb_000613_110 |
0.1022774753 |
- |
- |
PREDICTED: uncharacterized protein LOC105641540 [Jatropha curcas] |
| 16 |
Hb_009020_040 |
0.1031449603 |
- |
- |
PREDICTED: triacylglycerol lipase 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_028487_070 |
0.1037139993 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000261_050 |
0.1046394581 |
- |
- |
PREDICTED: probable magnesium transporter NIPA2 [Populus euphratica] |
| 19 |
Hb_000359_210 |
0.1058415685 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_023988_030 |
0.1064950087 |
- |
- |
PREDICTED: transmembrane protein 19 [Vitis vinifera] |