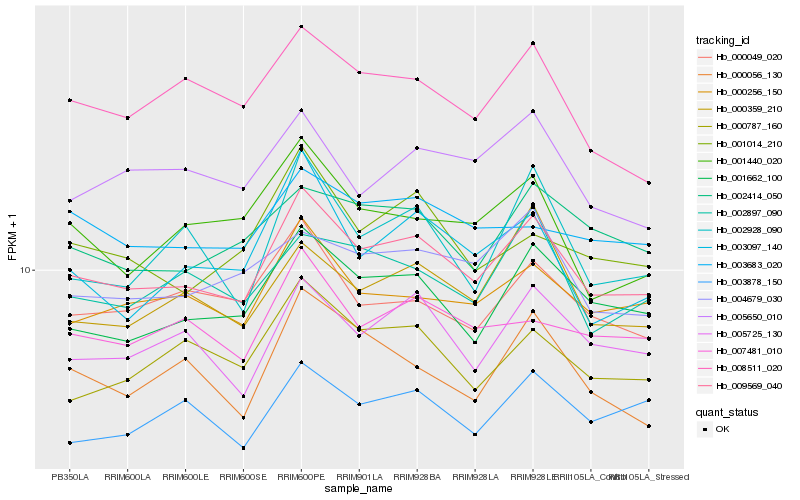
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009569_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635573 [Jatropha curcas] |
| 2 |
Hb_001662_100 |
0.0559567553 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like [Jatropha curcas] |
| 3 |
Hb_000256_150 |
0.0628845607 |
- |
- |
PREDICTED: trafficking protein particle complex II-specific subunit 120 homolog [Jatropha curcas] |
| 4 |
Hb_005725_130 |
0.0646301025 |
- |
- |
PREDICTED: uncharacterized protein KIAA0930 homolog isoform X2 [Jatropha curcas] |
| 5 |
Hb_000049_020 |
0.0716849948 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 6 |
Hb_007481_010 |
0.0718475841 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 7 |
Hb_002414_050 |
0.0735707514 |
- |
- |
hypothetical protein CICLE_v10000243mg [Citrus clementina] |
| 8 |
Hb_008511_020 |
0.0741409738 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 9 |
Hb_000787_160 |
0.0744509364 |
- |
- |
PREDICTED: GDP-mannose transporter GONST1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004679_030 |
0.0760231692 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |
| 11 |
Hb_002897_090 |
0.0776237552 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000359_210 |
0.0778124548 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001014_210 |
0.077895396 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 14 |
Hb_003878_150 |
0.0781556958 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 15 |
Hb_005650_010 |
0.0786465729 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_003097_140 |
0.0791959059 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 17 |
Hb_003683_020 |
0.0794430539 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 18 |
Hb_001440_020 |
0.0796316983 |
- |
- |
exocyst componenet sec8, putative [Ricinus communis] |
| 19 |
Hb_000056_130 |
0.0801362316 |
- |
- |
Sucrose Transporter 2C [Hevea brasiliensis subsp. brasiliensis] |
| 20 |
Hb_002928_090 |
0.0802116491 |
- |
- |
PREDICTED: metallocarboxypeptidase A-like protein TRV_02598 [Jatropha curcas] |