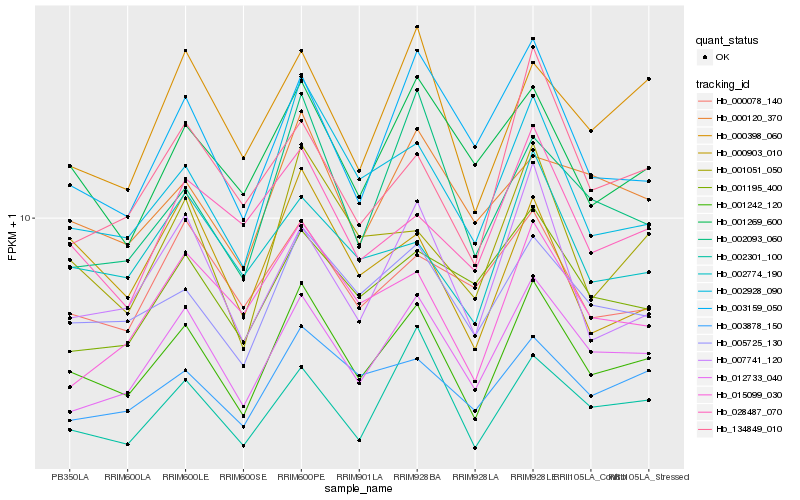
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001242_120 |
0.0 |
- |
- |
PREDICTED: carboxypeptidase D [Jatropha curcas] |
| 2 |
Hb_012733_040 |
0.0632302975 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 3 |
Hb_005725_130 |
0.0636744068 |
- |
- |
PREDICTED: uncharacterized protein KIAA0930 homolog isoform X2 [Jatropha curcas] |
| 4 |
Hb_002928_090 |
0.0676320386 |
- |
- |
PREDICTED: metallocarboxypeptidase A-like protein TRV_02598 [Jatropha curcas] |
| 5 |
Hb_002301_100 |
0.0756587048 |
- |
- |
hypothetical protein JCGZ_21479 [Jatropha curcas] |
| 6 |
Hb_001051_050 |
0.0797923723 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003878_150 |
0.0802016251 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 8 |
Hb_001269_600 |
0.083292379 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 9 |
Hb_000120_370 |
0.084950204 |
- |
- |
type 2 diacylglycerol acyltransferase [Ricinus communis] |
| 10 |
Hb_000398_060 |
0.0860964372 |
- |
- |
PREDICTED: survival of motor neuron-related-splicing factor 30 [Jatropha curcas] |
| 11 |
Hb_002774_190 |
0.0863292953 |
- |
- |
Endoplasmic reticulum-Golgi intermediate compartment protein, putative [Ricinus communis] |
| 12 |
Hb_007741_120 |
0.087364892 |
- |
- |
PREDICTED: phosphatidate cytidylyltransferase 4, chloroplastic [Jatropha curcas] |
| 13 |
Hb_003159_050 |
0.088512935 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 3, mitochondrial isoform X1 [Jatropha curcas] |
| 14 |
Hb_002093_060 |
0.0919304829 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 15 |
Hb_028487_070 |
0.0919941498 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_015099_030 |
0.0923517205 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 17 |
Hb_000903_010 |
0.0936524158 |
- |
- |
hypothetical protein JCGZ_23825 [Jatropha curcas] |
| 18 |
Hb_001195_400 |
0.0937997145 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 19 |
Hb_000078_140 |
0.0940078646 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 20 |
Hb_134849_010 |
0.0954792456 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |