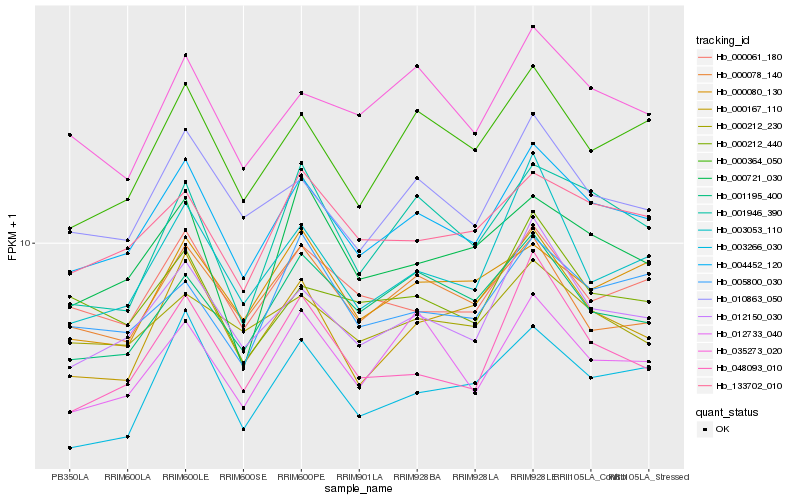
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004452_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639574 [Jatropha curcas] |
| 2 |
Hb_012150_030 |
0.0513754005 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 3 |
Hb_010863_050 |
0.058684384 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 4 |
Hb_133702_010 |
0.0613715127 |
- |
- |
PREDICTED: reticulon-4-interacting protein 1, mitochondrial isoform X1 [Jatropha curcas] |
| 5 |
Hb_000364_050 |
0.0625169039 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000061_180 |
0.0652182838 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 7 |
Hb_035273_020 |
0.0706220839 |
- |
- |
PREDICTED: amidase 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_012733_040 |
0.0722097908 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 9 |
Hb_003053_110 |
0.0724381009 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000212_230 |
0.0744673038 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 2 [Jatropha curcas] |
| 11 |
Hb_001195_400 |
0.0747900202 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 12 |
Hb_000080_130 |
0.0753595142 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 13 |
Hb_000212_440 |
0.0753823005 |
- |
- |
PREDICTED: riboflavin biosynthesis protein PYRR, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_003266_030 |
0.0761720607 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 15 |
Hb_005800_030 |
0.0763975984 |
- |
- |
PREDICTED: lysM and putative peptidoglycan-binding domain-containing protein 4-like [Jatropha curcas] |
| 16 |
Hb_000167_110 |
0.0778873721 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_001946_390 |
0.0781538196 |
- |
- |
PREDICTED: NADH-cytochrome b5 reductase-like protein [Jatropha curcas] |
| 18 |
Hb_000078_140 |
0.0792463555 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 19 |
Hb_048093_010 |
0.0801917793 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_000721_030 |
0.0819208201 |
rubber biosynthesis |
Gene Name: Mevalonate diphosphate decarboxylase |
mevalonate diphosphate decarboxylase [Hevea brasiliensis] |