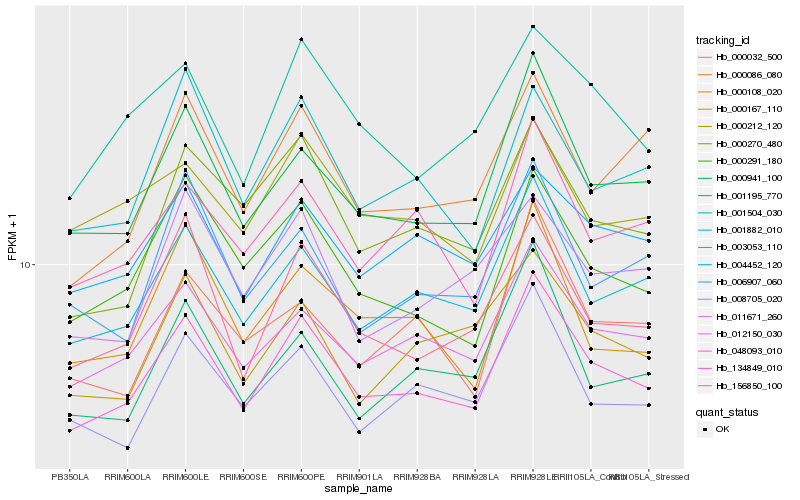
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_048093_010 |
0.0 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 2 |
Hb_000941_100 |
0.0610960874 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 3 |
Hb_003053_110 |
0.0622378862 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_012150_030 |
0.0632649394 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 5 |
Hb_156850_100 |
0.0742033305 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 6 |
Hb_000291_180 |
0.0766844354 |
- |
- |
PREDICTED: meiotically up-regulated gene 185 protein [Jatropha curcas] |
| 7 |
Hb_134849_010 |
0.0781646935 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 8 |
Hb_008705_020 |
0.0795002163 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 9 |
Hb_004452_120 |
0.0801917793 |
- |
- |
PREDICTED: uncharacterized protein LOC105639574 [Jatropha curcas] |
| 10 |
Hb_001504_030 |
0.0807422114 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 6 [Jatropha curcas] |
| 11 |
Hb_000270_480 |
0.0833629833 |
- |
- |
Mannose-1-phosphate guanyltransferase alpha [Gossypium arboreum] |
| 12 |
Hb_001195_770 |
0.0868382292 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 13 |
Hb_000108_020 |
0.0874015459 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 14 |
Hb_001882_010 |
0.0875632985 |
- |
- |
- |
| 15 |
Hb_000167_110 |
0.0886438258 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_000086_080 |
0.089792811 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000212_120 |
0.0920346127 |
- |
- |
hypothetical protein JCGZ_18070 [Jatropha curcas] |
| 18 |
Hb_000032_500 |
0.0939975446 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 19 |
Hb_006907_060 |
0.0945310284 |
- |
- |
Alternative oxidase 4, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_011671_260 |
0.0948515399 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X1 [Jatropha curcas] |