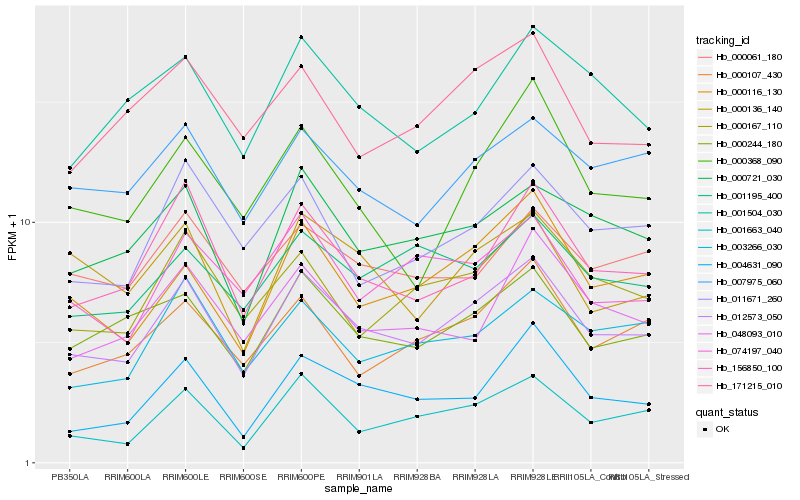
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012573_050 |
0.0 |
- |
- |
choline/ethanolamine kinase family protein [Populus trichocarpa] |
| 2 |
Hb_000244_180 |
0.0825292879 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_156850_100 |
0.0827425412 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 4 |
Hb_000721_030 |
0.0860930264 |
rubber biosynthesis |
Gene Name: Mevalonate diphosphate decarboxylase |
mevalonate diphosphate decarboxylase [Hevea brasiliensis] |
| 5 |
Hb_000116_130 |
0.0877497272 |
- |
- |
PREDICTED: aspartic proteinase Asp1 [Jatropha curcas] |
| 6 |
Hb_000167_110 |
0.0919430557 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_007975_060 |
0.0936528261 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000107_430 |
0.09403163 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 9 |
Hb_074197_040 |
0.097465101 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 10 |
Hb_011671_260 |
0.0975614642 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001663_040 |
0.0977539555 |
- |
- |
hypothetical protein B456_007G162000 [Gossypium raimondii] |
| 12 |
Hb_003266_030 |
0.0982382551 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 13 |
Hb_000061_180 |
0.0994198409 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 14 |
Hb_171215_010 |
0.0999410744 |
- |
- |
vesicle-associated membrane protein, putative [Ricinus communis] |
| 15 |
Hb_004631_090 |
0.1008782823 |
- |
- |
PREDICTED: probable glucan 1,3-beta-glucosidase A [Jatropha curcas] |
| 16 |
Hb_001195_400 |
0.1016317224 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 17 |
Hb_048093_010 |
0.1018602518 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_001504_030 |
0.1022740268 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 6 [Jatropha curcas] |
| 19 |
Hb_000368_090 |
0.1027294676 |
- |
- |
TPA: hypothetical protein ZEAMMB73_442906 [Zea mays] |
| 20 |
Hb_000136_140 |
0.1029353995 |
- |
- |
PREDICTED: uncharacterized protein LOC105648960 [Jatropha curcas] |