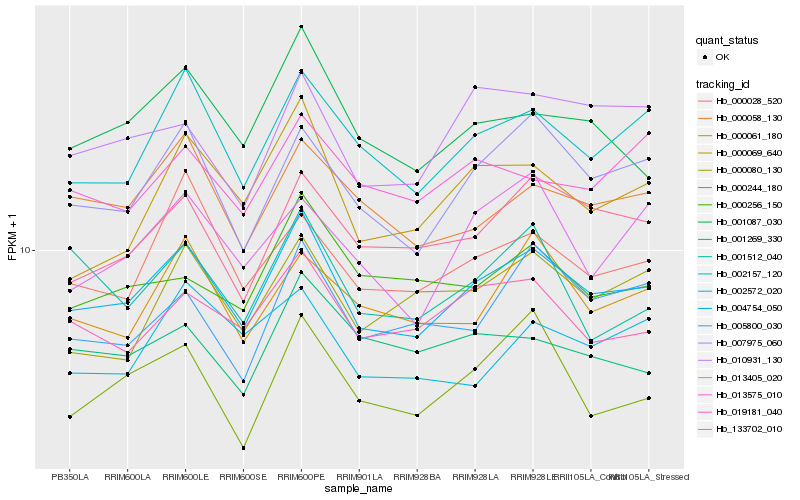
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004754_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104433925 [Eucalyptus grandis] |
| 2 |
Hb_007975_060 |
0.0622178795 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002157_120 |
0.0637433619 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_019181_040 |
0.0717296252 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 5 |
Hb_133702_010 |
0.0717365159 |
- |
- |
PREDICTED: reticulon-4-interacting protein 1, mitochondrial isoform X1 [Jatropha curcas] |
| 6 |
Hb_001087_030 |
0.0727500452 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit [Jatropha curcas] |
| 7 |
Hb_000058_130 |
0.0744749696 |
- |
- |
PREDICTED: uncharacterized protein LOC105640884 [Jatropha curcas] |
| 8 |
Hb_000028_520 |
0.0783275425 |
- |
- |
hypothetical protein L484_025125 [Morus notabilis] |
| 9 |
Hb_000069_640 |
0.0792449076 |
- |
- |
PREDICTED: tubulin gamma-1 chain [Vitis vinifera] |
| 10 |
Hb_000061_180 |
0.081027355 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 11 |
Hb_010931_130 |
0.0820204509 |
- |
- |
PREDICTED: mitogen-activated protein kinase 20 [Jatropha curcas] |
| 12 |
Hb_005800_030 |
0.082182381 |
- |
- |
PREDICTED: lysM and putative peptidoglycan-binding domain-containing protein 4-like [Jatropha curcas] |
| 13 |
Hb_000244_180 |
0.0843324372 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000080_130 |
0.0850357289 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 15 |
Hb_000256_150 |
0.0852221502 |
- |
- |
PREDICTED: trafficking protein particle complex II-specific subunit 120 homolog [Jatropha curcas] |
| 16 |
Hb_013575_010 |
0.0864047329 |
- |
- |
Hepatocyte growth factor-regulated tyrosine kinase substrate, putative [Ricinus communis] |
| 17 |
Hb_001512_040 |
0.0878809104 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |
| 18 |
Hb_001269_330 |
0.0880600238 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 19 |
Hb_013405_020 |
0.0891267622 |
- |
- |
- |
| 20 |
Hb_002572_020 |
0.0892231666 |
transcription factor |
TF Family: SBP |
hypothetical protein POPTR_0005s28010g [Populus trichocarpa] |