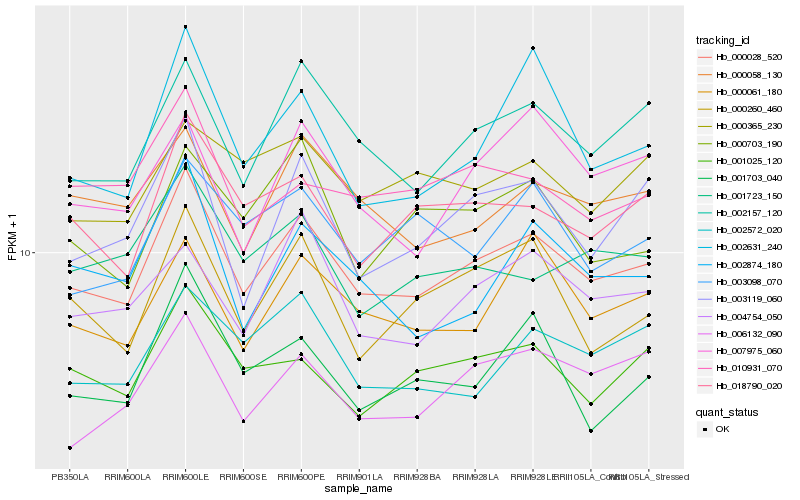
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000028_520 |
0.0 |
- |
- |
hypothetical protein L484_025125 [Morus notabilis] |
| 2 |
Hb_002157_120 |
0.0596268084 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_018790_020 |
0.0674856613 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 4 |
Hb_000061_180 |
0.0718254204 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 5 |
Hb_001025_120 |
0.0722590011 |
transcription factor |
TF Family: Jumonji |
PREDICTED: jmjC domain-containing protein 4 [Jatropha curcas] |
| 6 |
Hb_000058_130 |
0.0731205889 |
- |
- |
PREDICTED: uncharacterized protein LOC105640884 [Jatropha curcas] |
| 7 |
Hb_003119_060 |
0.0743131191 |
- |
- |
PREDICTED: dicarboxylate transporter 2.1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000260_460 |
0.0752578532 |
- |
- |
Ethanolamine-phosphate cytidylyltransferase, putative [Ricinus communis] |
| 9 |
Hb_006132_090 |
0.0762631417 |
- |
- |
PREDICTED: origin of replication complex subunit 5 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002874_180 |
0.0767727553 |
- |
- |
PREDICTED: protein AIG2-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_002631_240 |
0.0781110294 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 12 |
Hb_004754_050 |
0.0783275425 |
- |
- |
PREDICTED: uncharacterized protein LOC104433925 [Eucalyptus grandis] |
| 13 |
Hb_001703_040 |
0.0784783171 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002572_020 |
0.0787495551 |
transcription factor |
TF Family: SBP |
hypothetical protein POPTR_0005s28010g [Populus trichocarpa] |
| 15 |
Hb_001723_150 |
0.0792564796 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_010931_070 |
0.0798982033 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003098_070 |
0.082044029 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 18 |
Hb_000365_230 |
0.0825793169 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |
| 19 |
Hb_000703_190 |
0.0826065413 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_007975_060 |
0.0830829764 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X2 [Jatropha curcas] |