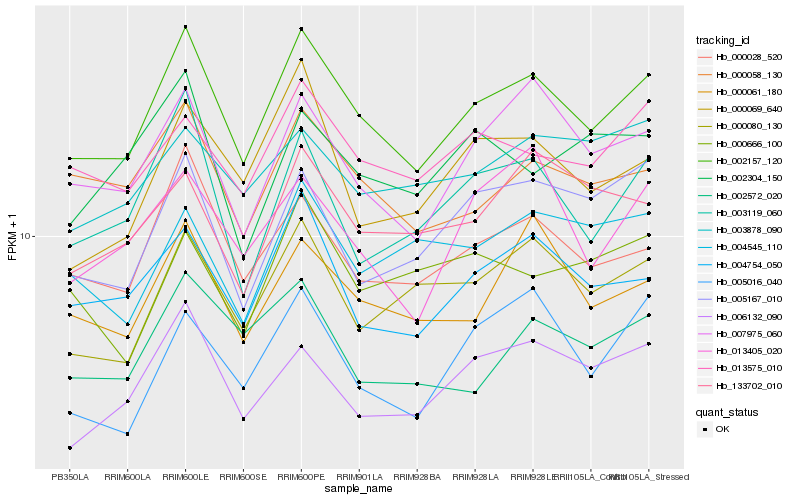
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002157_120 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000028_520 |
0.0596268084 |
- |
- |
hypothetical protein L484_025125 [Morus notabilis] |
| 3 |
Hb_004754_050 |
0.0637433619 |
- |
- |
PREDICTED: uncharacterized protein LOC104433925 [Eucalyptus grandis] |
| 4 |
Hb_007975_060 |
0.0647956987 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000058_130 |
0.0683441146 |
- |
- |
PREDICTED: uncharacterized protein LOC105640884 [Jatropha curcas] |
| 6 |
Hb_000061_180 |
0.0705849511 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 7 |
Hb_013575_010 |
0.0708951724 |
- |
- |
Hepatocyte growth factor-regulated tyrosine kinase substrate, putative [Ricinus communis] |
| 8 |
Hb_006132_090 |
0.0728053101 |
- |
- |
PREDICTED: origin of replication complex subunit 5 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000080_130 |
0.073134362 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 10 |
Hb_003119_060 |
0.0757703809 |
- |
- |
PREDICTED: dicarboxylate transporter 2.1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000069_640 |
0.0801317094 |
- |
- |
PREDICTED: tubulin gamma-1 chain [Vitis vinifera] |
| 12 |
Hb_002572_020 |
0.0804542363 |
transcription factor |
TF Family: SBP |
hypothetical protein POPTR_0005s28010g [Populus trichocarpa] |
| 13 |
Hb_013405_020 |
0.0809427261 |
- |
- |
- |
| 14 |
Hb_133702_010 |
0.0811903786 |
- |
- |
PREDICTED: reticulon-4-interacting protein 1, mitochondrial isoform X1 [Jatropha curcas] |
| 15 |
Hb_005167_010 |
0.0823603333 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X1 [Jatropha curcas] |
| 16 |
Hb_005016_040 |
0.0827372432 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor-like protein DPB isoform X2 [Jatropha curcas] |
| 17 |
Hb_003878_090 |
0.0846194438 |
- |
- |
PREDICTED: polynucleotide 3'-phosphatase ZDP [Jatropha curcas] |
| 18 |
Hb_004545_110 |
0.0864321064 |
- |
- |
DHHC-type zinc finger family protein isoform 1 [Theobroma cacao] |
| 19 |
Hb_000666_100 |
0.0898034127 |
- |
- |
Actin-related protein 2/3 complex subunit 2 isoform 2 [Theobroma cacao] |
| 20 |
Hb_002304_150 |
0.090282693 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |