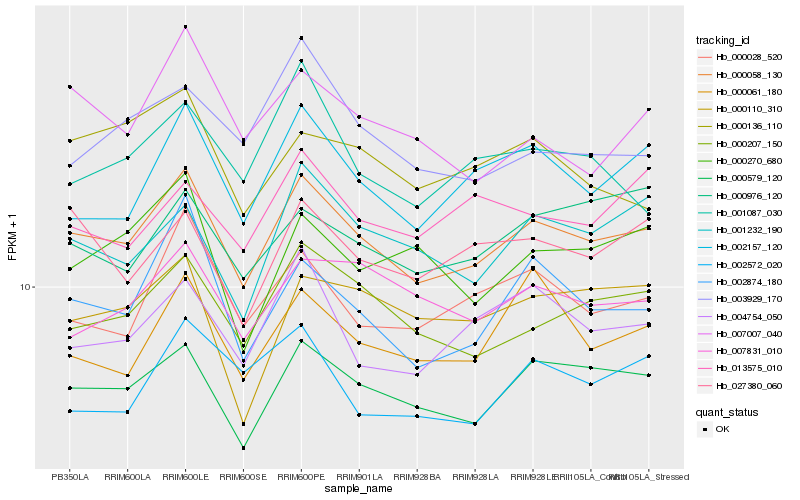
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000058_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640884 [Jatropha curcas] |
| 2 |
Hb_000579_120 |
0.0506294554 |
- |
- |
PREDICTED: uncharacterized protein LOC105633845 [Jatropha curcas] |
| 3 |
Hb_000207_150 |
0.0619130721 |
- |
- |
PREDICTED: uncharacterized membrane protein At4g09580 [Jatropha curcas] |
| 4 |
Hb_002157_120 |
0.0683441146 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000028_520 |
0.0731205889 |
- |
- |
hypothetical protein L484_025125 [Morus notabilis] |
| 6 |
Hb_001232_190 |
0.0735835871 |
- |
- |
PREDICTED: uncharacterized protein LOC105639761 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002874_180 |
0.0738636216 |
- |
- |
PREDICTED: protein AIG2-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_004754_050 |
0.0744749696 |
- |
- |
PREDICTED: uncharacterized protein LOC104433925 [Eucalyptus grandis] |
| 9 |
Hb_027380_060 |
0.076814745 |
- |
- |
PREDICTED: putative ALA-interacting subunit 2 [Jatropha curcas] |
| 10 |
Hb_007831_010 |
0.0768512451 |
- |
- |
PREDICTED: oligoribonuclease [Vitis vinifera] |
| 11 |
Hb_000061_180 |
0.0789303198 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 12 |
Hb_000976_120 |
0.0813133138 |
- |
- |
PREDICTED: DNA polymerase zeta processivity subunit [Jatropha curcas] |
| 13 |
Hb_007007_040 |
0.0818207917 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 14 |
Hb_000110_310 |
0.0834620627 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 4 of pyruvate dehydrogenase complex |
Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_013575_010 |
0.0838093073 |
- |
- |
Hepatocyte growth factor-regulated tyrosine kinase substrate, putative [Ricinus communis] |
| 16 |
Hb_000270_680 |
0.0844821733 |
- |
- |
hypothetical protein POPTR_0015s08420g [Populus trichocarpa] |
| 17 |
Hb_002572_020 |
0.0847775872 |
transcription factor |
TF Family: SBP |
hypothetical protein POPTR_0005s28010g [Populus trichocarpa] |
| 18 |
Hb_003929_170 |
0.0855652072 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 19 |
Hb_000136_110 |
0.0859688092 |
- |
- |
hypothetical protein B456_011G031000, partial [Gossypium raimondii] |
| 20 |
Hb_001087_030 |
0.0861656418 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit [Jatropha curcas] |