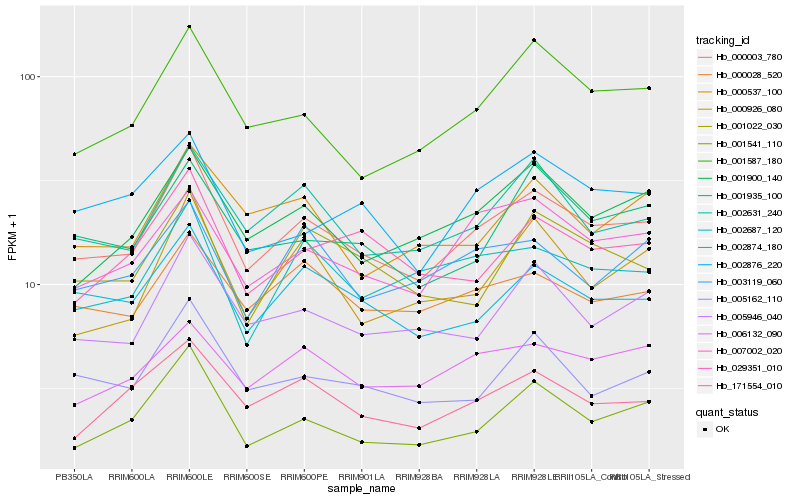
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_780 |
0.0 |
- |
- |
hexokinase [Manihot esculenta] |
| 2 |
Hb_002631_240 |
0.0791359858 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 3 |
Hb_005946_040 |
0.0814950261 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002874_180 |
0.0815245576 |
- |
- |
PREDICTED: protein AIG2-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_007002_020 |
0.0827594092 |
- |
- |
PREDICTED: uncharacterized protein LOC105642604 [Jatropha curcas] |
| 6 |
Hb_001900_140 |
0.0840396069 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001587_180 |
0.0849834155 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |
| 8 |
Hb_000028_520 |
0.0850172969 |
- |
- |
hypothetical protein L484_025125 [Morus notabilis] |
| 9 |
Hb_001541_110 |
0.0885279836 |
- |
- |
PREDICTED: sufE-like protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 10 |
Hb_005162_110 |
0.0887818268 |
- |
- |
PREDICTED: uncharacterized protein LOC105648430 [Jatropha curcas] |
| 11 |
Hb_171554_010 |
0.0890202184 |
- |
- |
PREDICTED: uncharacterized protein LOC105634071 [Jatropha curcas] |
| 12 |
Hb_006132_090 |
0.0898599868 |
- |
- |
PREDICTED: origin of replication complex subunit 5 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001935_100 |
0.0924596578 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 14 |
Hb_002687_120 |
0.0942332532 |
- |
- |
PREDICTED: lipoyl synthase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_029351_010 |
0.0948252128 |
- |
- |
PREDICTED: HD domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001022_030 |
0.0969042337 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 17 |
Hb_002876_220 |
0.0970819064 |
- |
- |
PREDICTED: uncharacterized protein LOC105633932 [Jatropha curcas] |
| 18 |
Hb_000926_080 |
0.097713706 |
- |
- |
PREDICTED: paraspeckle component 1 [Jatropha curcas] |
| 19 |
Hb_000537_100 |
0.0984225095 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 20 |
Hb_003119_060 |
0.0992578729 |
- |
- |
PREDICTED: dicarboxylate transporter 2.1, chloroplastic [Jatropha curcas] |