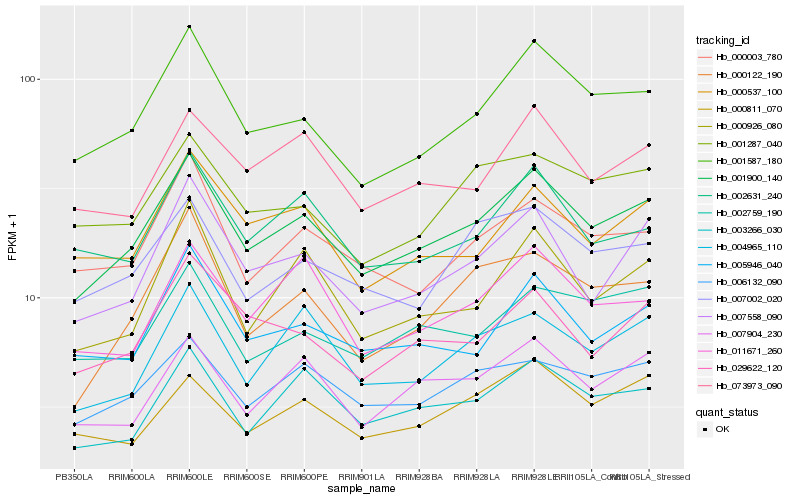
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001900_140 |
0.0 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001587_180 |
0.0644125213 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |
| 3 |
Hb_006132_090 |
0.0717889344 |
- |
- |
PREDICTED: origin of replication complex subunit 5 isoform X1 [Jatropha curcas] |
| 4 |
Hb_007558_090 |
0.0731980245 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN8, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_003266_030 |
0.0752782589 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 6 |
Hb_007904_230 |
0.0766759241 |
- |
- |
PREDICTED: D-cysteine desulfhydrase 2, mitochondrial [Jatropha curcas] |
| 7 |
Hb_005946_040 |
0.0808004159 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 8 |
Hb_007002_020 |
0.0811798705 |
- |
- |
PREDICTED: uncharacterized protein LOC105642604 [Jatropha curcas] |
| 9 |
Hb_073973_090 |
0.0821107276 |
- |
- |
Aminotransferase ybdL, putative [Ricinus communis] |
| 10 |
Hb_000926_080 |
0.0822490968 |
- |
- |
PREDICTED: paraspeckle component 1 [Jatropha curcas] |
| 11 |
Hb_004965_110 |
0.0823061315 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 12 |
Hb_000122_190 |
0.083306147 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000003_780 |
0.0840396069 |
- |
- |
hexokinase [Manihot esculenta] |
| 14 |
Hb_000537_100 |
0.0841884552 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 15 |
Hb_029622_120 |
0.0843548444 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2 [Jatropha curcas] |
| 16 |
Hb_001287_040 |
0.0850372579 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 17 |
Hb_002759_190 |
0.0858740326 |
- |
- |
PREDICTED: protein NLRC3 [Jatropha curcas] |
| 18 |
Hb_000811_070 |
0.0858964391 |
- |
- |
PREDICTED: uncharacterized protein LOC105650695 [Jatropha curcas] |
| 19 |
Hb_011671_260 |
0.0861524873 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002631_240 |
0.0864046012 |
- |
- |
JHL17M24.3 [Jatropha curcas] |