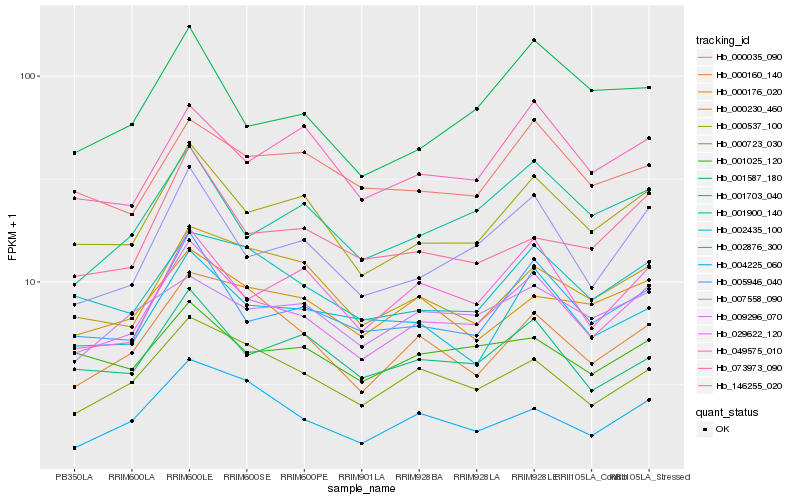
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029622_120 |
0.0 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2 [Jatropha curcas] |
| 2 |
Hb_000537_100 |
0.0586205951 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 3 |
Hb_007558_090 |
0.0668274472 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN8, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_005946_040 |
0.0711505329 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000723_030 |
0.0786179323 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor-like protein DPB isoform X1 [Jatropha curcas] |
| 6 |
Hb_001900_140 |
0.0843548444 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002435_100 |
0.0855202864 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF217 [Jatropha curcas] |
| 8 |
Hb_049575_010 |
0.0858885777 |
- |
- |
hypothetical protein POPTR_0001s15330g [Populus trichocarpa] |
| 9 |
Hb_001703_040 |
0.0877938817 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002876_300 |
0.0941039341 |
- |
- |
PREDICTED: histidinol-phosphate aminotransferase, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_000176_020 |
0.094544065 |
- |
- |
PREDICTED: nuclear pore complex protein NUP43 [Jatropha curcas] |
| 12 |
Hb_000230_460 |
0.0945683937 |
- |
- |
Speckle-type POZ protein, putative [Ricinus communis] |
| 13 |
Hb_004225_060 |
0.0947674926 |
- |
- |
hypothetical protein CISIN_1g027511mg [Citrus sinensis] |
| 14 |
Hb_073973_090 |
0.095443165 |
- |
- |
Aminotransferase ybdL, putative [Ricinus communis] |
| 15 |
Hb_000160_140 |
0.0958738168 |
- |
- |
PREDICTED: ELL-associated factor 2-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_146255_020 |
0.0967491731 |
- |
- |
PREDICTED: chromatin structure-remodeling complex protein BSH [Jatropha curcas] |
| 17 |
Hb_001025_120 |
0.0973251645 |
transcription factor |
TF Family: Jumonji |
PREDICTED: jmjC domain-containing protein 4 [Jatropha curcas] |
| 18 |
Hb_009296_070 |
0.0981408637 |
- |
- |
PREDICTED: cyclin-dependent kinase D-3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000035_090 |
0.1001199821 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 20 |
Hb_001587_180 |
0.1007320111 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |