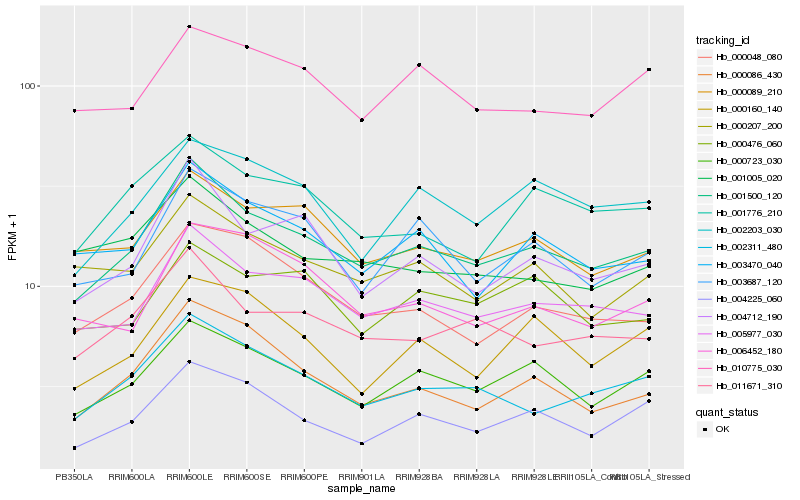
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001500_120 |
0.0 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 2 |
Hb_005977_030 |
0.0593573528 |
- |
- |
PREDICTED: monothiol glutaredoxin-S10-like [Jatropha curcas] |
| 3 |
Hb_000723_030 |
0.0699940812 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor-like protein DPB isoform X1 [Jatropha curcas] |
| 4 |
Hb_004712_190 |
0.0727944555 |
- |
- |
PREDICTED: uncharacterized protein LOC105633105 [Jatropha curcas] |
| 5 |
Hb_002311_480 |
0.0750735761 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_011671_310 |
0.0759315677 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2 [Jatropha curcas] |
| 7 |
Hb_003470_040 |
0.0822418932 |
- |
- |
PREDICTED: 2-oxoisovalerate dehydrogenase subunit beta 1, mitochondrial [Jatropha curcas] |
| 8 |
Hb_004225_060 |
0.0826340123 |
- |
- |
hypothetical protein CISIN_1g027511mg [Citrus sinensis] |
| 9 |
Hb_000089_210 |
0.0841477577 |
- |
- |
unknown [Medicago truncatula] |
| 10 |
Hb_003687_120 |
0.0860486144 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |
| 11 |
Hb_000086_430 |
0.0862563955 |
- |
- |
hypothetical protein JCGZ_17649 [Jatropha curcas] |
| 12 |
Hb_000048_080 |
0.0874820576 |
- |
- |
PREDICTED: cyclin-dependent kinase D-3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000207_200 |
0.0909495455 |
- |
- |
PREDICTED: histidinol-phosphate aminotransferase, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_001776_210 |
0.0920312152 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 15 |
Hb_002203_030 |
0.0924382863 |
transcription factor |
TF Family: NF-YB |
PREDICTED: protein Dr1 homolog isoform X2 [Jatropha curcas] |
| 16 |
Hb_006452_180 |
0.0998042428 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 17 |
Hb_001005_020 |
0.1008579333 |
- |
- |
PREDICTED: uncharacterized protein LOC105630212 [Jatropha curcas] |
| 18 |
Hb_010775_030 |
0.1011020041 |
- |
- |
PREDICTED: 60S ribosomal protein L4 [Vitis vinifera] |
| 19 |
Hb_000476_060 |
0.1021441259 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_000160_140 |
0.1025252431 |
- |
- |
PREDICTED: ELL-associated factor 2-like isoform X1 [Jatropha curcas] |